RAD21 mutations cause a human cohesinopathy
- PMID: 22633399
- PMCID: PMC3370273
- DOI: 10.1016/j.ajhg.2012.04.019
RAD21 mutations cause a human cohesinopathy
Abstract
The evolutionarily conserved cohesin complex was originally described for its role in regulating sister-chromatid cohesion during mitosis and meiosis. Cohesin and its regulatory proteins have been implicated in several human developmental disorders, including Cornelia de Lange (CdLS) and Roberts syndromes. Here we show that human mutations in the integral cohesin structural protein RAD21 result in a congenital phenotype consistent with a "cohesinopathy." Children with RAD21 mutations display growth retardation, minor skeletal anomalies, and facial features that overlap findings in individuals with CdLS. Notably, unlike children with mutations in NIPBL, SMC1A, or SMC3, these individuals have much milder cognitive impairment than those with classical CdLS. Mechanistically, these mutations act at the RAD21 interface with the other cohesin proteins STAG2 and SMC1A, impair cellular DNA damage response, and disrupt transcription in a zebrafish model. Our data suggest that, compared to loss-of-function mutations, dominant missense mutations result in more severe functional defects and cause worse structural and cognitive clinical findings. These results underscore the essential role of RAD21 in eukaryotes and emphasize the need for further understanding of the role of cohesin in human development.
Copyright © 2012 The American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures

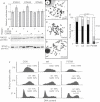

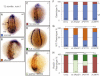
References
-
- Nasmyth K., Haering C.H. Cohesin: Its roles and mechanisms. Annu. Rev. Genet. 2009;43:525–558. - PubMed
-
- Watrin E., Schleiffer A., Tanaka K., Eisenhaber F., Nasmyth K., Peters J.M. Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr. Biol. 2006;16:863–874. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

