Tailoring enzymes involved in the biosynthesis of angucyclines contain latent context-dependent catalytic activities
- PMID: 22633416
- PMCID: PMC3361699
- DOI: 10.1016/j.chembiol.2012.04.010
Tailoring enzymes involved in the biosynthesis of angucyclines contain latent context-dependent catalytic activities
Abstract
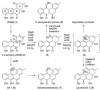
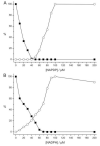
Comparison of homologous angucycline modification enzymes from five closely related Streptomyces pathways (pga, cab, jad, urd, lan) allowed us to deduce the biosynthetic steps responsible for the three alternative outcomes: gaudimycin C, dehydrorabelomycin, and 11-deoxylandomycinone. The C-12b-hydroxylated urdamycin and gaudimycin metabolites appear to be the ancestral representatives from which landomycins and jadomysins have evolved as a result of functional divergence of the ketoreductase LanV and hydroxylase JadH, respectively. Specifically, LanV has acquired affinity for an earlier biosynthetic intermediate resulting in a switch in biosynthetic order and lack of hydroxyls at C-4a and C-12b, whereas in JadH, C-4a/C-12b dehydration has evolved into an independent secondary function replacing C-12b hydroxylation. Importantly, the study reveals that many of the modification enzymes carry several alternative, hidden, or ancestral catalytic functions, which are strictly dependent on the biosynthetic context.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures



Similar articles
-
Structural and functional analysis of angucycline C-6 ketoreductase LanV involved in landomycin biosynthesis.Biochemistry. 2013 Aug 6;52(31):5304-14. doi: 10.1021/bi400712q. Epub 2013 Jul 24. Biochemistry. 2013. PMID: 23848284
-
Flavoprotein hydroxylase PgaE catalyzes two consecutive oxygen-dependent tailoring reactions in angucycline biosynthesis.Biochemistry. 2011 Jun 21;50(24):5535-43. doi: 10.1021/bi200600k. Epub 2011 May 27. Biochemistry. 2011. PMID: 21595438
-
Artificial reconstruction of two cryptic angucycline antibiotic biosynthetic pathways.Chembiochem. 2007 Sep 3;8(13):1577-84. doi: 10.1002/cbic.200700140. Chembiochem. 2007. PMID: 17654627
-
The functional differentiation of the post-PKS tailoring oxygenases contributed to the chemical diversities of atypical angucyclines.Synth Syst Biotechnol. 2018 Nov 14;3(4):275-282. doi: 10.1016/j.synbio.2018.11.001. eCollection 2018 Dec. Synth Syst Biotechnol. 2018. PMID: 30533539 Free PMC article. Review.
-
Glycosyltransferases involved in the biosynthesis of biologically active natural products that contain oligosaccharides.Mol Biosyst. 2005 Jul;1(2):117-26. doi: 10.1039/b503215f. Epub 2005 Jun 23. Mol Biosyst. 2005. PMID: 16880973 Review.
Cited by
-
Himalaquinones A-G, Angucyclinone-Derived Metabolites Produced by the Himalayan Isolate Streptomyces sp. PU-MM59.J Nat Prod. 2021 Jul 23;84(7):1930-1940. doi: 10.1021/acs.jnatprod.1c00192. Epub 2021 Jun 25. J Nat Prod. 2021. PMID: 34170698 Free PMC article.
-
Characterization of the Biosynthetic Gene Cluster and Shunt Products Yields Insights into the Biosynthesis of Balmoralmycin.Appl Environ Microbiol. 2022 Dec 13;88(23):e0120822. doi: 10.1128/aem.01208-22. Epub 2022 Nov 9. Appl Environ Microbiol. 2022. PMID: 36350133 Free PMC article.
-
Divergent evolution of an atypical S-adenosyl-l-methionine-dependent monooxygenase involved in anthracycline biosynthesis.Proc Natl Acad Sci U S A. 2015 Aug 11;112(32):9866-71. doi: 10.1073/pnas.1501765112. Epub 2015 Jul 27. Proc Natl Acad Sci U S A. 2015. PMID: 26216966 Free PMC article.
-
Heterologous expression of the cryptic mdk gene cluster and structural revision of maduralactomycin A.RSC Adv. 2023 Nov 22;13(48):34136-34144. doi: 10.1039/d3ra05931f. eCollection 2023 Nov 16. RSC Adv. 2023. PMID: 38019997 Free PMC article.
-
Functional and Structural Insights into a Novel Promiscuous Ketoreductase of the Lugdunomycin Biosynthetic Pathway.ACS Chem Biol. 2020 Sep 18;15(9):2529-2538. doi: 10.1021/acschembio.0c00564. Epub 2020 Sep 8. ACS Chem Biol. 2020. PMID: 32840360 Free PMC article.
References
-
- Antal N, Fiedler HP, Stackebrandt E, Beil W, Stroch K, Zeeck A. Retymicin, galtamycin B, saquayamycin Z and ribofuranosyllumichrome, novel secondary metabolites from Micromonospora sp. Tu 6368. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. (Tokyo) 2005;58:95–102. - PubMed
-
- Autschbach J, Ziegler T, van Gisbergen SJA, Baerends EJ. Chiroptical properties from time-dependent density functional theory. I. Circular dichroism spectra of organic molecule. J. Chem. Phys. 2002;116:6930–6940.
-
- Ayer SW, McInnes AG, Thibault P, Walter JA, Doull JL, Parnell T, Vining LC. Jadomycin, a novel 8H-benz[b]oxazolo[3,2-f]phenanthridine antibiotic from from Streptomyces venezuelae ISP5230. Tetrahedron Lett. 1991;32:6301–6304.
-
- Chen Y, Fan K, He Y, Xu X, Peng Y, Yu T, Jia C, Yang K. Characterization of JadH as an FAD- and NAD(P)H-dependent bifunctional hydroxylase/dehydrase in jadomycin biosynthesis. ChemBioChem. 2010;11:1055–1060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

