Isolation of primitive endoderm, mesoderm, vascular endothelial and trophoblast progenitors from human pluripotent stem cells
- PMID: 22634564
- PMCID: PMC3672406
- DOI: 10.1038/nbt.2239
Isolation of primitive endoderm, mesoderm, vascular endothelial and trophoblast progenitors from human pluripotent stem cells
Abstract
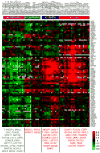
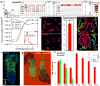
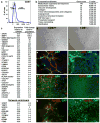
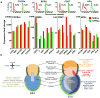
To identify early populations of committed progenitors derived from human embryonic stem cells (hESCs), we screened self-renewing, BMP4-treated and retinoic acid-treated cultures with >400 antibodies recognizing cell-surface antigens. Sorting of >30 subpopulations followed by transcriptional analysis of developmental genes identified four distinct candidate progenitor groups. Subsets detected in self-renewing cultures, including CXCR4(+) cells, expressed primitive endoderm genes. Expression of Cxcr4 in primitive endoderm was confirmed in visceral endoderm of mouse embryos. BMP4-induced progenitors exhibited gene signatures of mesoderm, trophoblast and vascular endothelium, suggesting correspondence to gastrulation-stage primitive streak, chorion and allantois precursors, respectively. Functional studies in vitro and in vivo confirmed that ROR2(+) cells produce mesoderm progeny, APA(+) cells generate syncytiotrophoblasts and CD87(+) cells give rise to vasculature. The same progenitor classes emerged during the differentiation of human induced pluripotent stem cells (hiPSCs). These markers and progenitors provide tools for purifying human tissue-regenerating progenitors and for studying the commitment of pluripotent stem cells to lineage progenitors.
Conflict of interest statement
The authors declare no competing financial interests
Figures



Comment in
-
Taming stem cell heterogeneity.Nat Methods. 2012 Jul;9(7):645. doi: 10.1038/nmeth.2094. Nat Methods. 2012. PMID: 22930832 No abstract available.
References
-
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. - PubMed
-
- Xu RH, et al. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol. 2002;20:1261–1264. - PubMed
-
- D’Amour KA, et al. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. - PubMed
-
- Yasunaga M, et al. Induction and monitoring of definitive and visceral endoderm differentiation of mouse ES cells. Nat Biotechnol. 2005;23:1542–1550. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

