Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk
- PMID: 22634755
- PMCID: PMC4747430
- DOI: 10.1038/ng.2293
Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk
Abstract
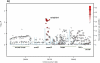
We performed a meta-analysis of five genome-wide association studies to identify common variants influencing colorectal cancer (CRC) risk comprising 8,682 cases and 9,649 controls. Replication analysis was performed in case-control sets totaling 21,096 cases and 19,555 controls. We identified three new CRC risk loci at 6p21 (rs1321311, near CDKN1A; P = 1.14 × 10(-10)), 11q13.4 (rs3824999, intronic to POLD3; P = 3.65 × 10(-10)) and Xp22.2 (rs5934683, near SHROOM2; P = 7.30 × 10(-10)) This brings the number of independent loci associated with CRC risk to 20 and provides further insight into the genetic architecture of inherited susceptibility to CRC.
Figures


Comment in
-
Meta-analysis of several GWAS sets yields additional genetic susceptibility variants for colorectal cancer: first X-linked component identified.Gastroenterology. 2012 Dec;143(6):1684-5. doi: 10.1053/j.gastro.2012.10.008. Epub 2012 Oct 13. Gastroenterology. 2012. PMID: 23073136 No abstract available.
References
-
- Lichtenstein P, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. - PubMed
-
- Aaltonen L, Johns L, Jarvinen H, Mecklin JP, Houlston R. Explaining the familial colorectal cancer risk associated with mismatch repair (MMR)-deficient and MMR-stable tumors. Clin Cancer Res. 2007;13:356–61. - PubMed
-
- Lubbe SJ, Webb EL, Chandler IP, Houlston RS. Implications of familial colorectal cancer risk profiles and microsatellite instability status. J Clin Oncol. 2009;27:2238–44. - PubMed
-
- Tomlinson IP, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 05-0001/AICR_/Worldwide Cancer Research/United Kingdom
- U01 CA074799/CA/NCI NIH HHS/United States
- 12076/CRUK_/Cancer Research UK/United Kingdom
- BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_U122861325/MRC_/Medical Research Council/United Kingdom
- C10195/A12996/CRUK_/Cancer Research UK/United Kingdom
- U01 CA097735/CA/NCI NIH HHS/United States
- CA-95-011/CA/NCI NIH HHS/United States
- U01 CA074783/CA/NCI NIH HHS/United States
- 10589/CRUK_/Cancer Research UK/United Kingdom
- C1298/A8362/CRUK_/Cancer Research UK/United Kingdom
- 10124/CRUK_/Cancer Research UK/United Kingdom
- MC_U127527198/MRC_/Medical Research Council/United Kingdom
- U01CA122839/CA/NCI NIH HHS/United States
- C490/A10124/CRUK_/Cancer Research UK/United Kingdom
- U01 CA074794/CA/NCI NIH HHS/United States
- C31250/A10107/CRUK_/Cancer Research UK/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- 090532/Z/09/Z/WT_/Wellcome Trust/United Kingdom
- C10314/A4886/CRUK_/Cancer Research UK/United Kingdom
- C348/A12076/CRUK_/Cancer Research UK/United Kingdom
- G1002033/MRC_/Medical Research Council/United Kingdom
- U01 CA122839/CA/NCI NIH HHS/United States
- 076113/C/04/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

