Cellular characterization of the primosome and rep helicase in processing and restoration of replication following arrest by UV-induced DNA damage in Escherichia coli
- PMID: 22636770
- PMCID: PMC3416541
- DOI: 10.1128/JB.00290-12
Cellular characterization of the primosome and rep helicase in processing and restoration of replication following arrest by UV-induced DNA damage in Escherichia coli
Abstract
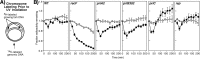
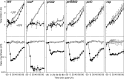
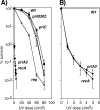
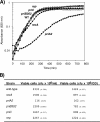
Following arrest by UV-induced DNA damage, replication is restored through a sequence of steps that involve partial resection of the nascent DNA by RecJ and RecQ, branch migration and processing of the fork DNA surrounding the lesion by RecA and RecF-O-R, and resumption of DNA synthesis once the blocking lesion has been repaired or bypassed. In vitro, the primosomal proteins (PriA, PriB, and PriC) and Rep are capable of initiating replication from synthetic DNA fork structures, and they have been proposed to catalyze these events when replication is disrupted by certain impediments in vivo. Here, we characterized the role that PriA, PriB, PriC, and Rep have in processing and restoring replication forks following arrest by UV-induced DNA damage. We show that the partial degradation and processing of the arrested replication fork occurs normally in both rep and primosome mutants. In each mutant, the nascent degradation ceases and DNA synthesis initially resumes in a timely manner, but the recovery then stalls in the absence of PriA, PriB, or Rep. The results demonstrate a role for the primosome and Rep helicase in overcoming replication forks arrested by UV-induced damage in vivo and suggest that these proteins are required for the stability and efficiency of the replisome when DNA synthesis resumes but not to initiate de novo replication downstream of the lesion.
Figures


Similar articles
-
Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli.Proc Natl Acad Sci U S A. 2006 Jun 13;103(24):9154-9. doi: 10.1073/pnas.0600785103. Epub 2006 Jun 5. Proc Natl Acad Sci U S A. 2006. PMID: 16754873 Free PMC article.
-
RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli.Mol Gen Genet. 1999 Oct;262(3):543-51. doi: 10.1007/s004380051116. Mol Gen Genet. 1999. PMID: 10589843
-
RecO acts with RecF and RecR to protect and maintain replication forks blocked by UV-induced DNA damage in Escherichia coli.J Biol Chem. 2004 Jan 30;279(5):3492-6. doi: 10.1074/jbc.M311012200. Epub 2003 Nov 18. J Biol Chem. 2004. PMID: 14625283
-
Dissecting the functional role of PriA protein-catalysed primosome assembly in Escherichia coli DNA replication.Mol Microbiol. 1991 Dec;5(12):2869-73. doi: 10.1111/j.1365-2958.1991.tb01846.x. Mol Microbiol. 1991. PMID: 1667219 Review.
-
Participation of recombination proteins in rescue of arrested replication forks in UV-irradiated Escherichia coli need not involve recombination.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8196-202. doi: 10.1073/pnas.121008898. Proc Natl Acad Sci U S A. 2001. PMID: 11459953 Free PMC article. Review.
Cited by
-
Functions that Protect Escherichia coli from Tightly Bound DNA-Protein Complexes Created by Mutant EcoRII Methyltransferase.PLoS One. 2015 May 19;10(5):e0128092. doi: 10.1371/journal.pone.0128092. eCollection 2015. PLoS One. 2015. PMID: 25993347 Free PMC article.
-
Mechanisms of bacterial DNA replication restart.Nucleic Acids Res. 2018 Jan 25;46(2):504-519. doi: 10.1093/nar/gkx1203. Nucleic Acids Res. 2018. PMID: 29202195 Free PMC article. Review.
-
Plasma-sensitive Escherichia coli mutants reveal plasma resistance mechanisms.J R Soc Interface. 2019 Mar 29;16(152):20180846. doi: 10.1098/rsif.2018.0846. J R Soc Interface. 2019. PMID: 30913981 Free PMC article.
-
Homologous recombination as a replication fork escort: fork-protection and recovery.Biomolecules. 2012 Dec 27;3(1):39-71. doi: 10.3390/biom3010039. Biomolecules. 2012. PMID: 24970156 Free PMC article.
-
Regulation of Rep helicase unwinding by an auto-inhibitory subdomain.Nucleic Acids Res. 2019 Mar 18;47(5):2523-2532. doi: 10.1093/nar/gkz023. Nucleic Acids Res. 2019. PMID: 30690484 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

