Genome sequencing of a genetically tractable Pyrococcus furiosus strain reveals a highly dynamic genome
- PMID: 22636780
- PMCID: PMC3416535
- DOI: 10.1128/JB.00439-12
Genome sequencing of a genetically tractable Pyrococcus furiosus strain reveals a highly dynamic genome
Abstract
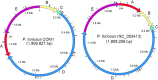
The model archaeon Pyrococcus furiosus grows optimally near 100°C on carbohydrates and peptides. Its genome sequence (NCBI) was determined 12 years ago. A genetically tractable strain, COM1, was very recently reported, and here we describe its genome sequence. Of 1,909,827 bp in size, it is 1,571 bp longer (0.1%) than the reference NCBI sequence. The COM1 genome contains numerous chromosomal rearrangements, deletions, and single base changes. COM1 also has 45 full or partial insertion sequences (ISs) compared to 35 in the reference NCBI strain, and these have resulted in the direct deletion or insertional inactivation of 13 genes. Another seven genes were affected by chromosomal deletions and are predicted to be nonfunctional. In addition, the amino acid sequences of another 102 of the 2,134 predicted gene products are different in COM1. These changes potentially impact various cellular functions, including carbohydrate, peptide, and nucleotide metabolism; DNA repair; CRISPR-associated defense; transcriptional regulation; membrane transport; and growth at 72°C. For example, the IS-mediated inactivation of riboflavin synthase in COM1 resulted in a riboflavin requirement for growth. Nevertheless, COM1 grew on cellobiose, malto-oligosaccharides, and peptides in complex and minimal media at 98 and 72°C to the same extent as did both its parent strain and a new culture collection strain (DSMZ 3638). This was in spite of COM1 lacking several metabolic enzymes, including nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase and beta-glucosidase. The P. furiosus genome is therefore of high plasticity, and the availability of the COM1 sequence will be critical for the future studies of this model hyperthermophile.
Figures


References
-
- Albers SV, et al. 2009. SulfoSYS (Sulfolobus Systems Biology): towards a silicon cell model for the central carbohydrate metabolism of the archaeon Sulfolobus solfataricus under temperature variation. Biochem. Soc. Trans. 37:58–64 - PubMed
-
- Atomi H. 2005. Recent progress towards the application of hyperthermophiles and their enzymes. Curr. Opin. Chem. Biol. 9:166–173 - PubMed
-
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 - PubMed
-
- Brunner NA, Brinkmann H, Siebers B, Hensel R. 1998. NAD+-dependent glyceraldehyde-3-phosphate dehydrogenase from Thermoproteus tenax. The first identified archaeal member of the aldehyde dehydrogenase superfamily is a glycolytic enzyme with unusual regulatory properties. J. Biol. Chem. 273:6149–6156 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

