Research resource: A genome-wide study identifies potential new target genes for POU1F1
- PMID: 22638072
- PMCID: PMC5416979
- DOI: 10.1210/me.2011-1308
Research resource: A genome-wide study identifies potential new target genes for POU1F1
Abstract
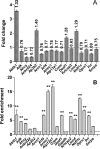
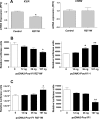
The pituitary transcription factor POU1F1 is required for the differentiation of lactotrope, thyrotrope, and somatotrope cells. Its expression is maintained in the adult and is crucial for the expression of prolactin, GH, and TSHβ-subunit. Different studies indicated that POU1F1 could also have other functions in these cells. The identification of new targets of this factor could be useful to obtain a better understanding of these functions. To address this question we combined data obtained from expression microarrays and from chromatin immunoprecipitation (ChIP)-chips. Gene expression microarray assays were used to detect genes that have their expression modified in somatolactotrope GH4C1 cells by the expression of a dominant-negative form of POU1F1, POU1F1(R271W), and led to the identification of 1346 such genes. ChIP-chip experiments were performed from mouse pituitaries and identified 1671 POU1F1-binding sites in gene-promoter regions. Intersecting the gene expression and the ChIP-chip data yielded 121 potential new direct targets. The initial set of 1346 genes identified using the microarrays, as well as the 121 potential new direct targets, were analyzed with DAVID bioinformatics resource for gene ontology term enrichment and cluster. This analysis revealed enrichment in different terms related to protein synthesis and transport, to apoptosis, and to cell division. The present study represents an integrative genome-wide approach to identify new target genes of POU1F1 and downstream networks controlled by this factor.
Figures
References
-
- Bodner M , Castrillo JL , Theill LE , Deerinck T , Ellisman M , Karin M. 1988. The pituitary-specific transcription factor GHF-1 is a homeobox-containing protein. Cell 55:505–518 - PubMed
-
- Ingraham HA , Chen RP , Mangalam HJ , Elsholtz HP , Flynn SE , Lin CR , Simmons DM , Swanson L , Rosenfeld MG. 1988. A tissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell 55:519–529 - PubMed
-
- Li S , Crenshaw EB , Rawson EJ , Simmons DM , Swanson LW , Rosenfeld MG. 1990. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature 347:528–533 - PubMed
-
- Radovick S , Nations M , Du Y , Berg LA , Weintraub BD , Wondisford FE. 1992. A mutation in the POU-homeodomain of Pit-1 responsible for combined pituitary hormone deficiency. Science 257:1115–1118 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

