Myh7b/miR-499 gene expression is transcriptionally regulated by MRFs and Eos
- PMID: 22638570
- PMCID: PMC3424578
- DOI: 10.1093/nar/gks466
Myh7b/miR-499 gene expression is transcriptionally regulated by MRFs and Eos
Abstract
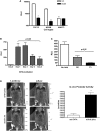
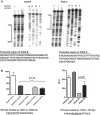
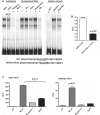
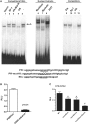
The sarcomeric myosin gene, Myh7b, encodes an intronic microRNA, miR-499, which regulates cardiac and skeletal muscle biology, yet little is known about its transcriptional regulation. To identify the transcription factors involved in regulating Myh7b/miR-499 gene expression, we have mapped the transcriptional start sites and identified an upstream 6.2 kb region of the mouse Myh7b gene whose activity mimics the expression pattern of the endogenous Myh7b gene both in vitro and in vivo. Through promoter deletion analysis, we have mapped a distal E-box element and a proximal Ikaros site that are essential for Myh7b promoter activity in muscle cells. We show that the myogenic regulatory factors, MyoD, Myf5 and Myogenin, bind to the E-box, while a lymphoid transcription factor, Ikaros 4 (Eos), binds to the Ikaros motif. Further, we show that through physical interaction, MyoD and Eos form an active transcriptional complex on the chromatin to regulate the expression of the endogenous Myh7b/miR-499 gene in muscle cells. We also provide the first evidence that Eos can regulate expression of additional myosin genes (Myosin 1 and β-Myosin) via the miR-499/Sox6 pathway. Therefore, our results indicate a novel role for Eos in the regulation of the myofiber gene program.
Figures





Similar articles
-
Developmental expression and cardiac transcriptional regulation of Myh7b, a third myosin heavy chain in the vertebrate heart.Cytoskeleton (Hoboken). 2012 May;69(5):324-35. doi: 10.1002/cm.21029. Epub 2012 Apr 30. Cytoskeleton (Hoboken). 2012. PMID: 22422726 Free PMC article.
-
Activation of the beta myosin heavy chain promoter by MEF-2D, MyoD, p300, and the calcineurin/NFATc1 pathway.J Cell Physiol. 2007 Apr;211(1):138-48. doi: 10.1002/jcp.20916. J Cell Physiol. 2007. PMID: 17111365
-
A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance.Dev Cell. 2009 Nov;17(5):662-73. doi: 10.1016/j.devcel.2009.10.013. Dev Cell. 2009. PMID: 19922871 Free PMC article.
-
Nonproductive Splicing Prevents Expression of MYH7b Protein in the Mammalian Heart.J Am Heart Assoc. 2021 Jul 20;10(14):e020965. doi: 10.1161/JAHA.121.020965. Epub 2021 Jul 6. J Am Heart Assoc. 2021. PMID: 34227390 Free PMC article.
-
Regulation and functions of myogenic regulatory factors in lower vertebrates.Comp Biochem Physiol B Biochem Mol Biol. 2001 Aug;130(1):1-12. doi: 10.1016/s1096-4959(01)00412-2. Comp Biochem Physiol B Biochem Mol Biol. 2001. PMID: 11470439 Review.
Cited by
-
Effect of fetal hypothyroidism on MyomiR network and its target gene expression profiles in heart of offspring rats.Mol Cell Biochem. 2017 Dec;436(1-2):179-187. doi: 10.1007/s11010-017-3089-7. Epub 2017 Jun 28. Mol Cell Biochem. 2017. PMID: 28660410
-
Identification of MyoD-Responsive Transcripts Reveals a Novel Long Non-coding RNA (lncRNA-AK143003) that Negatively Regulates Myoblast Differentiation.Sci Rep. 2017 Jun 6;7(1):2828. doi: 10.1038/s41598-017-03071-7. Sci Rep. 2017. PMID: 28588232 Free PMC article.
-
Hyperhomocysteinemia associated skeletal muscle weakness involves mitochondrial dysfunction and epigenetic modifications.Biochim Biophys Acta. 2015 May;1852(5):732-41. doi: 10.1016/j.bbadis.2015.01.008. Epub 2015 Jan 20. Biochim Biophys Acta. 2015. PMID: 25615794 Free PMC article.
-
Regulation of microRNA function in somatic stem cell proliferation and differentiation.Nat Rev Mol Cell Biol. 2014 Sep;15(9):565-76. doi: 10.1038/nrm3854. Epub 2014 Aug 13. Nat Rev Mol Cell Biol. 2014. PMID: 25118717 Free PMC article. Review.
-
Evolution of the myosin heavy chain gene MYH14 and its intronic microRNA miR-499: muscle-specific miR-499 expression persists in the absence of the ancestral host gene.BMC Evol Biol. 2013 Jul 6;13:142. doi: 10.1186/1471-2148-13-142. BMC Evol Biol. 2013. PMID: 24059862 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

