Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily
- PMID: 22638584
- PMCID: PMC3424549
- DOI: 10.1093/nar/gks382
Sequence, structure and functional diversity of PD-(D/E)XK phosphodiesterase superfamily
Abstract
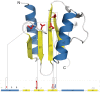
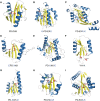
Proteins belonging to PD-(D/E)XK phosphodiesterases constitute a functionally diverse superfamily with representatives involved in replication, restriction, DNA repair and tRNA-intron splicing. Their malfunction in humans triggers severe diseases, such as Fanconi anemia and Xeroderma pigmentosum. To date there have been several attempts to identify and classify new PD-(D/E)KK phosphodiesterases using remote homology detection methods. Such efforts are complicated, because the superfamily exhibits extreme sequence and structural divergence. Using advanced homology detection methods supported with superfamily-wide domain architecture and horizontal gene transfer analyses, we provide a comprehensive reclassification of proteins containing a PD-(D/E)XK domain. The PD-(D/E)XK phosphodiesterases span over 21,900 proteins, which can be classified into 121 groups of various families. Eleven of them, including DUF4420, DUF3883, DUF4263, COG5482, COG1395, Tsp45I, HaeII, Eco47II, ScaI, HpaII and Replic_Relax, are newly assigned to the PD-(D/E)XK superfamily. Some groups of PD-(D/E)XK proteins are present in all domains of life, whereas others occur within small numbers of organisms. We observed multiple horizontal gene transfers even between human pathogenic bacteria or from Prokaryota to Eukaryota. Uncommon domain arrangements greatly elaborate the PD-(D/E)XK world. These include domain architectures suggesting regulatory roles in Eukaryotes, like stress sensing and cell-cycle regulation. Our results may inspire further experimental studies aimed at identification of exact biological functions, specific substrates and molecular mechanisms of reactions performed by these highly diverse proteins.
Figures
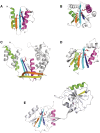
References
-
- Belfort M, Weiner A. Another bridge between kingdoms: tRNA splicing in archaea and eukaryotes. Cell. 1997;89:1003–1006. - PubMed
-
- Hickman AB, Li Y, Mathew SV, May EW, Craig NL, Dyda F. Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell. 2000;5:1025–1034. - PubMed
-
- Dahlroth SL, Gurmu D, Schmitzberger F, Engman H, Haas J, Erlandsen H, Nordlund P. Crystal structure of the shutoff and exonuclease protein from the oncogenic Kaposi's sarcoma-associated herpesvirus. FEBS J. 2009;276:6636–6645. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

