Linear motifs confer functional diversity onto splice variants
- PMID: 22638587
- PMCID: PMC3424572
- DOI: 10.1093/nar/gks442
Linear motifs confer functional diversity onto splice variants
Abstract
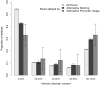
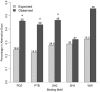
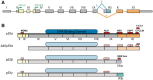
The pre-translational modification of messenger ribonucleic acids (mRNAs) by alternative promoter usage and alternative splicing is an important source of pleiotropy. Despite intensive efforts, our understanding of the functional implications of this dynamically created diversity is still incomplete. Using the available knowledge of interaction modules, particularly within intrinsically disordered regions (IDRs), we analysed the occurrences of protein modules within alternative exons. We find that regions removed or included by pre-translational variation are enriched in linear motifs suggesting that the removal or inclusion of exons containing these interaction modules is an important regulatory mechanism. In particular, we observe that PDZ-, PTB-, SH2- and WW-domain binding motifs are more likely to occur within alternative exons. We also determine that regions removed or included by alternative promoter usage are enriched in IDRs suggesting that protein isoform diversity is tightly coupled to the modulation of IDRs. This study, therefore, demonstrates that short linear motifs are key components for establishing protein diversity between splice variants.
Figures

References
-
- Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–177. - PubMed
-
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. - PubMed
-
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. - PubMed
-
- Resch A, Xing Y, Modrek B, Gorlick M, Riley R, Lee C. Assessing the impact of alternative splicing on domain interactions in the human proteome. J. Proteome. Res. 2004;3:76–83. - PubMed

