Conserved and diversified gene families of monovalent cation/h(+) antiporters from algae to flowering plants
- PMID: 22639643
- PMCID: PMC3355601
- DOI: 10.3389/fpls.2012.00025
Conserved and diversified gene families of monovalent cation/h(+) antiporters from algae to flowering plants
Abstract
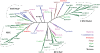
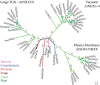
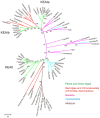
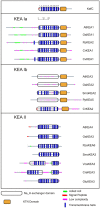
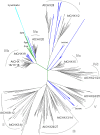
All organisms have evolved strategies to regulate ion and pH homeostasis in response to developmental and environmental cues. One strategy is mediated by monovalent cation-proton antiporters (CPA) that are classified in two superfamilies. Many CPA1 genes from bacteria, fungi, metazoa, and plants have been functionally characterized; though roles of plant CPA2 genes encoding K(+)-efflux antiporter (KEA) and cation/H(+) exchanger (CHX) families are largely unknown. Phylogenetic analysis showed that three clades of the CPA1 Na(+)-H(+) exchanger (NHX) family have been conserved from single-celled algae to Arabidopsis. These are (i) plasma membrane-bound SOS1/AtNHX7 that share ancestry with prokaryote NhaP, (ii) endosomal AtNHX5/6 that is part of the eukaryote Intracellular-NHE clade, and (iii) a vacuolar NHX clade (AtNHX1-4) specific to plants. Early diversification of KEA genes possibly from an ancestral cyanobacterium gene is suggested by three types seen in all plants. Intriguingly, CHX genes diversified from three to four members in one subclade of early land plants to 28 genes in eight subclades of Arabidopsis. Homologs from Spirogyra or Physcomitrella share high similarity with AtCHX20, suggesting that guard cell-specific AtCHX20 and its closest relatives are founders of the family, and pollen-expressed CHX genes appeared later in monocots and early eudicots. AtCHX proteins mediate K(+) transport and pH homeostasis, and have been localized to intracellular and plasma membrane. Thus KEA genes are conserved from green algae to angiosperms, and their presence in red algae and secondary endosymbionts suggest a role in plastids. In contrast, AtNHX1-4 subtype evolved in plant cells to handle ion homeostasis of vacuoles. The great diversity of CHX genes in land plants compared to metazoa, fungi, or algae would imply a significant role of ion and pH homeostasis at dynamic endomembranes in the vegetative and reproductive success of flowering plants.
Keywords: cargo sorting; cation homeostasis; dynamic endomembrane; pH homeostasis; protein; secretory system.
Figures


References
-
- Abrahamsen M. S., Templeton T. J., Enomoto S., Abrahante J. E., Zhu G., Lancto C. A., Deng M., Liu C., Widmer G., Tzipori S., Buck G. A., Xu P., Bankier A. T., Dear P. H., Konfortov B. A., Spriggs H. F., Iyer L., Anantharaman V., Aravind L., Kapur V. (2004). Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304, 441–445 10.1126/science.1094786 - DOI - PubMed
-
- Adl S. M., Simpson A. G., Farmer M. A., Andersen R. A., Anderson O. R., Barta J. R., Bowser S. S., Brugerolle G., Fensome R. A., Fredericq S., James T. Y., Karpov S., Kugrens P., Krug J., Lane C. E., Lewis L. A., Lodge J., Lynn D. H., Mann D. G., McCourt R. M., Mendoza L., Moestrup O., Mozley-Standridge S. E., Nerad T. A., Shearer C. A., Smirnov A. V., Spiegel F. W., Taylor M. F. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–451 10.1111/j.1550-7408.2005.00053.x - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

