Annotation of Selaginella moellendorffii Major Intrinsic Proteins and the Evolution of the Protein Family in Terrestrial Plants
- PMID: 22639644
- PMCID: PMC3355642
- DOI: 10.3389/fpls.2012.00033
Annotation of Selaginella moellendorffii Major Intrinsic Proteins and the Evolution of the Protein Family in Terrestrial Plants
Abstract
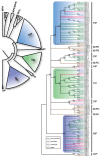
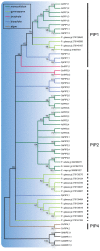
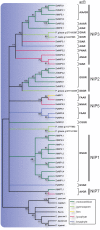
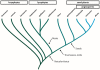
Major intrinsic proteins (MIPs) also called aquaporins form pores in membranes to facilitate the permeation of water and certain small polar solutes across membranes. MIPs are present in virtually every organism but are uniquely abundant in land plants. To elucidate the evolution and function of MIPs in terrestrial plants, the MIPs encoded in the genome of the spikemoss Selaginella moellendorffii were identified and analyzed. In total 19 MIPs were found in S. moellendorffii belonging to 6 of the 7 MIP subfamilies previously identified in the moss Physcomitrella patens. Only three of the MIPs were classified as members of the conserved water specific plasma membrane intrinsic protein (PIP) subfamily whereas almost half were found to belong to the diverse NOD26-like intrinsic protein (NIP) subfamily permeating various solutes. The small number of PIPs in S. moellendorffii is striking compared to all other land plants and no other species has more NIPs than PIPs. Similar to moss, S. moellendorffii only has one type of tonoplast intrinsic protein (TIP). Based on ESTs from non-angiosperms we conclude that the specialized groups of TIPs present in higher plants are not found in primitive vascular plants but evolved later in a common ancestor of seed plants. We also note that the silicic acid permeable NIP2 group that has been reported from angiosperms appears at the same time. We suggest that the expansion of the number MIP isoforms in higher plants is primarily associated with an increase in the different types of specialized tissues rather than the emergence of vascular tissue per se and that the loss of subfamilies has been possible due to a functional overlap between some subfamilies.
Keywords: AQP; GIP; HIP; SIP; XIP; phylogeny; water channels.
Figures


References
-
- Azad A. K., Yoshikawa N., Ishikawa T., Sawa Y., Shibata H. (2011). Substitution of a single amino acid residue in the aromatic/arginine selectivity filter alters the transport profiles of tonoplast aquaporin homologs. Biochim. Biophys. Acta 1818, 1–11 - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

