Modeling holo-ACP:DH and holo-ACP:KR complexes of modular polyketide synthases: a docking and molecular dynamics study
- PMID: 22639887
- PMCID: PMC3422181
- DOI: 10.1186/1472-6807-12-10
Modeling holo-ACP:DH and holo-ACP:KR complexes of modular polyketide synthases: a docking and molecular dynamics study
Abstract
Background: Modular polyketide synthases are multifunctional megasynthases which biosynthesize a variety of secondary metabolites using various combinations of dehydratase (DH), ketoreductase (KR) and enoyl-reductase (ER) domains. During the catalysis of various reductive steps these domains act on a substrate moiety which is covalently attached to the phosphopantetheine (P-pant) group of the holo-Acyl Carrier Protein (holo-ACP) domain, thus necessitating the formation of holo-ACP:DH and holo-ACP:KR complexes. Even though three dimensional structures are available for DH, KR and ACP domains, no structures are available for DH or KR domains in complex with ACP or substrate moieties. Since Ser of holo-ACP is covalently attached to a large phosphopantetheine group, obtaining complexes involving holo-ACP by standard protein-protein docking has been a difficult task.
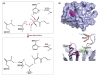
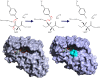
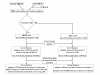
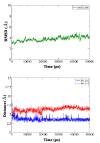
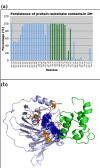
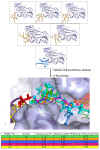
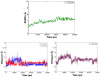
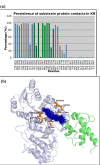
Results: We have modeled the holo-ACP:DH and holo-ACP:KR complexes for identifying specific residues on DH and KR domains which are involved in interaction with ACP, phosphopantetheine and substrate moiety. A novel combination of protein-protein and protein-ligand docking has been used to first model complexes involving apo-ACP and then dock the phosphopantetheine and substrate moieties using covalent connectivity between ACP, phosphopantetheine and substrate moiety as constraints. The holo-ACP:DH and holo-ACP:KR complexes obtained from docking have been further refined by restraint free explicit solvent MD simulations to incorporate effects of ligand and receptor flexibilities. The results from 50 ns MD simulations reveal that substrate enters into a deep tunnel in DH domain while in case of KR domain the substrate binds a shallow surface exposed cavity. Interestingly, in case of DH domain the predicted binding site overlapped with the binding site in the inhibitor bound crystal structure of FabZ, the DH domain from E.Coli FAS. In case of KR domain, the substrate binding site identified by our simulations was in proximity of the known stereo-specificity determining residues.
Conclusions: We have modeled the holo-ACP:DH and holo-ACP:KR complexes and identified the specific residues on DH and KR domains which are involved in interaction with ACP, phosphopantetheine and substrate moiety. Analysis of the conservation profile of binding pocket residues in homologous sequences of DH and KR domains indicated that, these results can also be extrapolated to reductive domains of other modular PKS clusters.
Figures

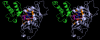


References
-
- Starcevic A, Zucko J, Simunkovic J, Long PF, Cullum J, Hranueli D. ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 2008;36(21):6882–6892. doi: 10.1093/nar/gkn685. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

