An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals
- PMID: 22640020
- PMCID: PMC3495650
- DOI: 10.1186/1471-2148-12-71
An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals
Abstract
Background: The APOBEC3 (A3) genes play a key role in innate antiviral defense in mammals by introducing directed mutations in the DNA. The human genome encodes for seven A3 genes, with multiple splice alternatives. Different A3 proteins display different substrate specificity, but the very basic question on how discerning self from non-self still remains unresolved. Further, the expression of A3 activity/ies shapes the way both viral and host genomes evolve.
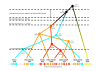
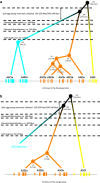
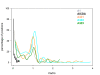
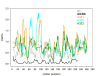
Results: We present here a detailed temporal analysis of the origin and expansion of the A3 repertoire in mammals. Our data support an evolutionary scenario where the genome of the mammalian ancestor encoded for at least one ancestral A3 gene, and where the genome of the ancestor of placental mammals (and possibly of the ancestor of all mammals) already encoded for an A3Z1-A3Z2-A3Z3 arrangement. Duplication events of the A3 genes have occurred independently in different lineages: humans, cats and horses. In all of them, gene duplication has resulted in changes in enzyme activity and/or substrate specificity, in a paradigmatic example of convergent adaptive evolution at the genomic level. Finally, our results show that evolutionary rates for the three A3Z1, A3Z2 and A3Z3 motifs have significantly decreased in the last 100 Mya. The analysis constitutes a textbook example of the evolution of a gene locus by duplication and sub/neofunctionalization in the context of virus-host arms race.
Conclusions: Our results provide a time framework for identifying ancestral and derived genomic arrangements in the APOBEC loci, and to date the expansion of this gene family for different lineages through time, as a response to changes in viral/retroviral/retrotransposon pressure.
Figures

References
-
- Tsuge M, Noguchi C, Akiyama R, Matsushita M, Kunihiro K, Tanaka S, Abe H, Mitsui F, Kitamura S, Hatakeyama T. et al.G to A hypermutation of TT virus. Virus Res. 2008;149(2):211–216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

