The NRF2-related interactome and regulome contain multifunctional proteins and fine-tuned autoregulatory loops
- PMID: 22641035
- PMCID: PMC7511993
- DOI: 10.1016/j.febslet.2012.05.016
The NRF2-related interactome and regulome contain multifunctional proteins and fine-tuned autoregulatory loops
Abstract
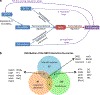
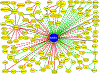
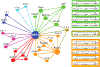
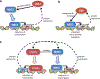
NRF2 is a well-known, master transcription factor (TF) of oxidative and xenobiotic stress responses. Recent studies uncovered an even wider regulatory role of NRF2 influencing carcinogenesis, inflammation and neurodegeneration. Prompted by these advances here we present a systems-level resource for NRF2 interactome and regulome that includes 289 protein-protein, 7469 TF-DNA and 85 miRNA interactions. As systems-level examples of NRF2-related signaling we identified regulatory loops of NRF2 interacting proteins (e.g., JNK1 and CBP) and a fine-tuned regulatory system, where 35 TFs regulated by NRF2 influence 63 miRNAs that down-regulate NRF2. The presented network and the uncovered regulatory loops may facilitate the development of efficient, NRF2-based therapeutic agents.
Copyright © 2012 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures
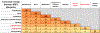
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

