GluN1 splice variant control of GluN1/GluN2D NMDA receptors
- PMID: 22641781
- PMCID: PMC3476637
- DOI: 10.1113/jphysiol.2012.234062
GluN1 splice variant control of GluN1/GluN2D NMDA receptors
Abstract
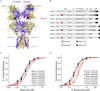
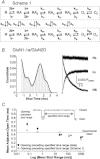
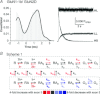
NMDA receptors are ionotropic glutamate receptors that mediate a slow, Ca2+-permeable component of excitatory synaptic transmission in the central nervous system. Recombinant GluN1-1a/GluN2D receptors are characterized by low channel open probability and prolonged deactivation time course following the removal of agonist. Here, we show that the deactivation time course, agonist potency, and single channel properties of GluN2D-containing NMDA receptors are modulated by alternative RNA splicing of GluN1. Our results demonstrate that GluN1 exon 5, which encodes a 21-amino-acid insert in the amino-terminal domain, is a key determinant of GluN1/GluN2D receptor function. GluN1-1b/GluN2D receptors, which contain the residues encoded by exon 5, deactivate with a dual exponential time course described by a τFAST of 410 ms and a τSLOW of 1100 ms. This time course is 3-fold more rapid than that for exon 5-lacking GluN1-1a/GluN2D, which deactivates with a τFAST of 1100 ms and a τSLOW of 3400 ms. Exon 5-containing NMDA receptors also have a two-fold higher open probability (0.037) than exon 5-lacking receptors (0.017). Furthermore, inclusion of exon 5-encoded residues within the GluN1-1b subunit decreases the potency for the endogenous agonist l-glutamate. Evaluation of receptor kinetics for NMDA receptors containing mutated GluN1-1b subunits and wild-type GluN2D identified residue Lys211 in GluN1-1b as a key determinant of exon 5 control of the deactivation time course and glutamate potency. Evaluation of a kinetic model of GluN1/GluN2D gating suggests that residues encoded by exon 5 influence several rate-limiting steps. These data demonstrate that the GluN1 subunit is a key determinant of the kinetic and pharmacological properties of GluN2D-containing NMDA receptors.
Figures




Comment in
-
The elusive roles of NMDA receptor amino-terminal domains.J Physiol. 2012 Nov 15;590(22):5561-2. doi: 10.1113/jphysiol.2012.243311. J Physiol. 2012. PMID: 23154855 Free PMC article. No abstract available.
References
-
- Banke TG, Traynelis SF. Activation of NR1/NR2B NMDA receptors. Nat Neurosci. 2003;6:144–152. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

