Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil- DNA glycosylase
- PMID: 22643842
- PMCID: PMC3388184
- DOI: 10.18632/oncotarget.503
Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil- DNA glycosylase
Abstract
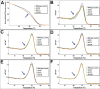
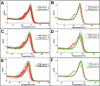
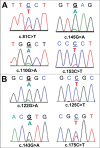
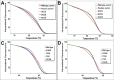
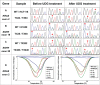
Non-reproducible sequence artefacts are frequently detected in DNA from formalinfixed and paraffin-embedded (FFPE) tissues. However, no rational strategy has been developed for reduction of sequence artefacts from FFPE DNA as the underlying causes of the artefacts are poorly understood. As cytosine deamination to uracil is a common form of DNA damage in ancient DNA, we set out to examine whether treatment of FFPE DNA with uracil-DNA glycosylase (UDG) would lead to the reduction of C>T (and G>A) sequence artefacts. Heteroduplex formation in high resolution melting (HRM)-based assays was used for the detection of sequence variants in FFPE DNA samples. A set of samples that gave false positive HRM results for screening for the E17K mutation in exon 4 of the AKT1 gene were chosen for analysis. Sequencing of these samples showed multiple non-reproducible C:G>T:A artefacts. Treatment of the FFPE DNA with UDG prior to PCR amplification led to a very marked reduction of the sequence artefacts as indicated by both HRM and sequencing analysis, indicating that uracil lesions are the major cause of sequence artefacts. Similar results were shown for the BRAF V600 region in the same sample set and EGFR exon 19 in another sample set. UDG treatment specifically suppressed the formation of artefacts in FFPE DNA as it did not affect the detection of true KRAS codon 12 and true EGFR exon 19 and 20 mutations. We conclude that uracil in FFPE DNA leads to a significant proportion of sequence artefacts. These can be minimised by a simple UDG pretreatment which can be readily carried out, in the same tube, as the PCR immediately prior to commencing thermal cycling. HRM is a convenient way of monitoring both the degree of damage and the effectiveness of the UDG treatment. These findings have immediate and important implications for cancer diagnostics where FFPE DNA is used as the primary genetic material for mutational studies guiding personalised medicine strategies and where simple effective strategies to detect mutations are required.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

