Caloric restriction-associated remodeling of rat white adipose tissue: effects on the growth hormone/insulin-like growth factor-1 axis, sterol regulatory element binding protein-1, and macrophage infiltration
- PMID: 22645024
- PMCID: PMC3705091
- DOI: 10.1007/s11357-012-9439-1
Caloric restriction-associated remodeling of rat white adipose tissue: effects on the growth hormone/insulin-like growth factor-1 axis, sterol regulatory element binding protein-1, and macrophage infiltration
Abstract
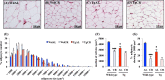
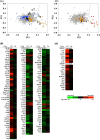
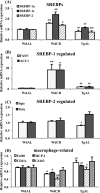
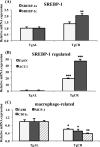
The role of the growth hormone (GH)-insulin-like growth factor (IGF)-1 axis in the lifelong caloric restriction (CR)-associated remodeling of white adipose tissue (WAT), adipocyte size, and gene expression profiles was explored in this study. We analyzed the WAT morphology of 6-7-month-old wild-type Wistar rats fed ad libitum (WdAL) or subjected to CR (WdCR), and of heterozygous transgenic dwarf rats bearing an anti-sense GH transgene fed ad libitum (TgAL) or subjected to CR (TgCR). Although less effective in TgAL, the adipocyte size was significantly reduced in WdCR compared with WdAL. This CR effect was blunted in Tg rats. We also used high-density oligonucleotide microarrays to examine the gene expression profile of WAT of WdAL, WdCR, and TgAL rats. The gene expression profile of WdCR, but not TgAL, differed greatly from that of WdAL. The gene clusters with the largest changes induced by CR but not by Tg were genes involved in lipid biosynthesis and inflammation, particularly sterol regulatory element binding proteins (SREBPs)-regulated and macrophage-related genes, respectively. Real-time reverse-transcription polymerase chain reaction analysis confirmed that the expression of SREBP-1 and its downstream targets was upregulated, whereas the macrophage-related genes were downregulated in WdCR, but not in TgAL. In addition, CR affected the gene expression profile of Tg rats similarly to wild-type rats. Our findings suggest that CR-associated remodeling of WAT, which involves SREBP-1-mediated transcriptional activation and suppression of macrophage infiltration, is regulated in a GH-IGF-1-independent manner.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
