Accurate de novo structure prediction of large transmembrane protein domains using fragment-assembly and correlated mutation analysis
- PMID: 22645369
- PMCID: PMC3386101
- DOI: 10.1073/pnas.1120036109
Accurate de novo structure prediction of large transmembrane protein domains using fragment-assembly and correlated mutation analysis
Abstract
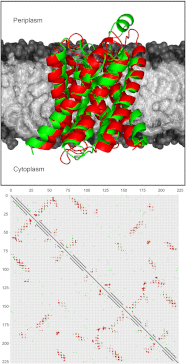
A new de novo protein structure prediction method for transmembrane proteins (FILM3) is described that is able to accurately predict the structures of large membrane proteins domains using an ensemble of two secondary structure prediction methods to guide fragment selection in combination with a scoring function based solely on correlated mutations detected in multiple sequence alignments. This approach has been validated by generating models for 28 membrane proteins with a diverse range of complex topologies and an average length of over 300 residues with results showing that TM-scores > 0.5 can be achieved in almost every case following refinement using MODELLER. In one of the most impressive results, a model of mitochondrial cytochrome c oxidase polypeptide I was obtained with a TM-score > 0.75 and an rmsd of only 5.7 Å over all 514 residues. These results suggest that FILM3 could be applicable to a wide range of transmembrane proteins of as-yet-unknown 3D structure given sufficient homologous sequences.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
All-atom 3D structure prediction of transmembrane β-barrel proteins from sequences.Proc Natl Acad Sci U S A. 2015 Apr 28;112(17):5413-8. doi: 10.1073/pnas.1419956112. Epub 2015 Apr 9. Proc Natl Acad Sci U S A. 2015. PMID: 25858953 Free PMC article.
-
Solvent and lipid accessibility prediction as a basis for model quality assessment in soluble and membrane proteins.Curr Protein Pept Sci. 2011 Sep;12(6):563-73. doi: 10.2174/138920311796957603. Curr Protein Pept Sci. 2011. PMID: 21787302
-
De novo membrane protein structure prediction.Methods Mol Biol. 2015;1215:331-50. doi: 10.1007/978-1-4939-1465-4_15. Methods Mol Biol. 2015. PMID: 25330970
-
Computational methods for protein secondary structure prediction using multiple sequence alignments.Curr Protein Pept Sci. 2000 Nov;1(3):273-301. doi: 10.2174/1389203003381324. Curr Protein Pept Sci. 2000. PMID: 12369910 Review.
-
Computational modeling of membrane proteins.Proteins. 2015 Jan;83(1):1-24. doi: 10.1002/prot.24703. Epub 2014 Nov 19. Proteins. 2015. PMID: 25355688 Free PMC article. Review.
Cited by
-
Protein structure refinement via molecular-dynamics simulations: What works and what does not?Proteins. 2016 Sep;84 Suppl 1(Suppl 1):282-92. doi: 10.1002/prot.24871. Epub 2015 Aug 17. Proteins. 2016. PMID: 26234208 Free PMC article.
-
SPARC: Structural properties associated with residue constraints.Comput Struct Biotechnol J. 2022 Apr 7;20:1702-1715. doi: 10.1016/j.csbj.2022.04.005. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35495120 Free PMC article.
-
Deep Analysis of Residue Constraints (DARC): identifying determinants of protein functional specificity.Sci Rep. 2020 Feb 3;10(1):1691. doi: 10.1038/s41598-019-55118-6. Sci Rep. 2020. PMID: 32015389 Free PMC article.
-
Coevolutionary signals across protein lineages help capture multiple protein conformations.Proc Natl Acad Sci U S A. 2013 Dec 17;110(51):20533-8. doi: 10.1073/pnas.1315625110. Epub 2013 Dec 2. Proc Natl Acad Sci U S A. 2013. PMID: 24297889 Free PMC article.
-
Network deconvolution as a general method to distinguish direct dependencies in networks.Nat Biotechnol. 2013 Aug;31(8):726-33. doi: 10.1038/nbt.2635. Epub 2013 Jul 14. Nat Biotechnol. 2013. PMID: 23851448 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

