The metabolite transporters of the plastid envelope: an update
- PMID: 22645538
- PMCID: PMC3355759
- DOI: 10.3389/fpls.2011.00050
The metabolite transporters of the plastid envelope: an update
Abstract
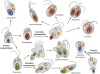
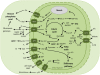
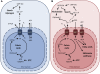
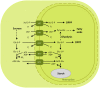
The engulfment of a photoautotrophic cyanobacterium by a primitive mitochondria-bearing eukaryote traces back to more than 1.2 billion years ago. This single endosymbiotic event not only provided the early petroalgae with the metabolic capacity to perform oxygenic photosynthesis, but also introduced a plethora of other metabolic routes ranging from fatty acids and amino acids biosynthesis, nitrogen and sulfur assimilation to secondary compounds synthesis. This implicated the integration and coordination of the newly acquired metabolic entity with the host metabolism. The interface between the host cytosol and the plastidic stroma became of crucial importance in sorting precursors and products between the plastid and other cellular compartments. The plastid envelope membranes fulfill different tasks: they perform important metabolic functions, as they are involved in the synthesis of carotenoids, chlorophylls, and galactolipids. In addition, since most genes of cyanobacterial origin have been transferred to the nucleus, plastidial proteins encoded by nuclear genes are post-translationally transported across the envelopes through the TIC-TOC import machinery. Most importantly, chloroplasts supply the photoautotrophic cell with photosynthates in form of reduced carbon. The innermost bilayer of the plastidic envelope represents the permeability barrier for the metabolites involved in the carbon cycle and is literally stuffed with transporter proteins facilitating their transfer. The intracellular metabolite transporters consist of polytopic proteins containing membrane spans usually in the number of four or more α-helices. Phylogenetic analyses revealed that connecting the plastid with the host metabolism was mainly a process driven by the host cell. In Arabidopsis, 58% of the metabolite transporters are of host origin, whereas only 12% are attributable to the cyanobacterial endosymbiont. This review focuses on the metabolite transporters of the inner envelope membrane of plastids, in particular the electrochemical potential-driven class of transporters. Recent advances in elucidating the plastidial complement of metabolite transporters are provided, with an update on phylogenetic relationship of selected proteins.
Keywords: endosymbiosis; envelope membrane; translocator.
Figures

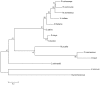
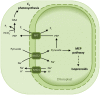
References
-
- Adl S. M., Simpson A. G., Farmer M. A., Andersen R. A., Anderson O. R., Barta J. R., Bowser S. S., Brugerolle G., Fensome R. A., Fredericq S., James T. Y., Karpov S., Kugrens P., Krug J., Lane C. E., Lewis L. A., Lodge J., Lynn D. H., Mann D. G., Mccourt R. M., Mendoza L., Moestrup O., Mozley-Standridge S. E., Nerad T. A., Shearer C. A., Smirnov A. V., Spiegel F. W., Taylor M. F. (2005). The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J. Eukaryot. Microbiol. 52, 399–45110.1111/j.1550-7408.2005.00053.x - DOI - PubMed
-
- Ardila F., Pozuetaromero J., Akazawa T. (1993). Adenylate uptake by proplastids from cultured-cells of tobacco (nicotiana-tabacum-L Cv-By2) indicates that an adenylate translocator is present in all types of plastid. Plant Cell Physiol. 34, 237–242
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

