Endoplasmic reticulum-associated degradation of glycoproteins in plants
- PMID: 22645596
- PMCID: PMC3355801
- DOI: 10.3389/fpls.2012.00067
Endoplasmic reticulum-associated degradation of glycoproteins in plants
Abstract
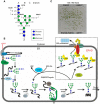
In all eukaryotes the endoplasmic reticulum (ER) has a central role in protein folding and maturation of secretory and membrane proteins. Upon translocation into the ER polypeptides are immediately subjected to folding and modifications involving the formation of disulfide bridges, assembly of subunits to multi-protein complexes, and glycosylation. During these processes incompletely folded, terminally misfolded, and unassembled proteins can accumulate which endanger the cellular homeostasis and subsequently the survival of cells and tissues. Consequently, organisms have developed a quality control system to cope with this problem and remove the unwanted protein load from the ER by a process collectively referred to as ER-associated degradation (ERAD) pathway. Recent studies in Arabidopsis have identified plant ERAD components involved in the degradation of aberrant proteins and evidence was provided for a specific role in abiotic stress tolerance. In this short review we discuss our current knowledge about this important cellular pathway.
Keywords: endoplasmic reticulum; protein degradation; protein glycosylation; protein quality control; ubiquitin–proteasome.
Figures
References
-
- Bernasconi R., Pertel T., Luban J., Molinari M. (2008). A dual task for the Xbp1-responsive OS-9 variants in the mammalian endoplasmic reticulum: inhibiting secretion of misfolded protein conformers and enhancing their disposal. J. Biol. Chem. 283, 16446–16454 10.1074/jbc.M802272200 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

