Discovery of catalytically active orthologues of the Parkinson's disease kinase PINK1: analysis of substrate specificity and impact of mutations
- PMID: 22645651
- PMCID: PMC3352081
- DOI: 10.1098/rsob.110012
Discovery of catalytically active orthologues of the Parkinson's disease kinase PINK1: analysis of substrate specificity and impact of mutations
Abstract
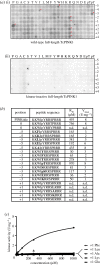
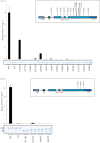
Missense mutations of the phosphatase and tensin homolog (PTEN)-induced kinase 1 (PINK1) gene cause autosomal-recessive Parkinson's disease. To date, little is known about the intrinsic catalytic properties of PINK1 since the human enzyme displays such low kinase activity in vitro. We have discovered that, in contrast to mammalian PINK1, insect orthologues of PINK1 we have investigated-namely Drosophila melanogaster (dPINK1), Tribolium castaneum (TcPINK1) and Pediculus humanus corporis (PhcPINK1)-are active as judged by their ability to phosphorylate the generic substrate myelin basic protein. We have exploited the most active orthologue, TcPINK1, to assess its substrate specificity and elaborated a peptide substrate (PINKtide, KKWIpYRRSPRRR) that can be employed to quantify PINK1 kinase activity. Analysis of PINKtide variants reveal that PINK1 phosphorylates serine or threonine, but not tyrosine, and we show that PINK1 exhibits a preference for a proline at the +1 position relative to the phosphorylation site. We have also, for the first time, been able to investigate the effect of Parkinson's disease-associated PINK1 missense mutations, and found that nearly all those located within the kinase domain, as well as the C-terminal non-catalytic region, markedly suppress kinase activity. This emphasizes the crucial importance of PINK1 kinase activity in preventing the development of Parkinson's disease. Our findings will aid future studies aimed at understanding how the activity of PINK1 is regulated and the identification of physiological substrates.
Keywords: biochemistry, Parkinson's disease, kinase.
Figures


References
-
- Valente EM, et al. 2004. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science 304, 1158–116010.1126/science.1096284 (doi:10.1126/science.1096284) - DOI - DOI - PubMed
-
- Abou-Sleiman PM, Muqit MM, Wood NW. 2006. Expanding insights of mitochondrial dysfunction in Parkinson's disease. Nat. Rev. Neurosci. 7, 207–21910.1038/nrn1868 (doi:10.1038/nrn1868) - DOI - DOI - PubMed
-
- Muqit MM, et al. 2006. Altered cleavage and localization of PINK1 to aggresomes in the presence of proteasomal stress. J. Neurochem. 98, 156–16910.1111/j.1471-4159.2006.03845.x (doi:10.1111/j.1471-4159.2006.03845.x) - DOI - DOI - PubMed
-
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR, Youle RJ. 2010. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298.10.1371/journal.pbio.1000298 (doi:10.1371/journal.pbio.1000298) - DOI - DOI - PMC - PubMed
-
- Matsuda N, et al. 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–22110.1083/jcb.200910140 (doi:10.1083/jcb.200910140) - DOI - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

