An iron-sulfur cluster loop motif in the Archaeoglobus fulgidus uracil-DNA glycosylase mediates efficient uracil recognition and removal
- PMID: 22646210
- PMCID: PMC3418143
- DOI: 10.1021/bi3000462
An iron-sulfur cluster loop motif in the Archaeoglobus fulgidus uracil-DNA glycosylase mediates efficient uracil recognition and removal
Abstract
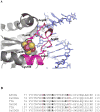
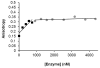
The family 4 uracil-DNA glycosylase from the hyperthermophilic organism Archaeoglobus fulgidus (AFUDG) is responsible for the removal of uracil in DNA as the first step in the base excision repair (BER) pathway. AFUDG contains a large solvent-exposed peptide region containing an α helix and loop anchored on each end via ligation of two cysteine thiolates to a [4Fe-4S](2+) cluster. We propose that this region plays a similar role in DNA damage recognition as a smaller iron-sulfur cluster loop (FCL) motif in the structurally unrelated BER glycosylases MutY and Endonuclease III and therefore refer to this region as the "pseudo-FCL" in AFUDG. In order to evaluate the importance of this region, three positively charged residues (Arg 86, Arg 91, Lys 100) and the anchoring Cys residues (Cys 85, Cys 101) within this motif were replaced with alanine, and the effects of these replacements on uracil excision in single- and double-stranded DNA were evaluated. These results show that this region participates and allows for efficient recognition and excision of uracil within DNA. Notably, R86A AFUDG exhibited reduced activity for uracil removal only within double-stranded DNA, suggesting an importance in duplex disruption and extrusion of the base as part of the excision process. In addition, mutation of the [4Fe-4S](2+) cluster cysteine ligands at the ends of the pseudo-FCL to alanine reduced the uracil excision efficiency, suggesting the importance of anchoring the loop via coordination to the cluster. In contrast, K100A AFUDG exhibited enhanced uracil excision activity, providing evidence for the importance of the loop conformation and flexibility. Taken together, the results herein provide evidence that the pseudo-FCL motif is involved in DNA binding and catalysis, particularly in duplex DNA contexts. This work underscores the requirement of an ensemble of interactions, both distant and in proximity to the damaged site, for accurate and efficient uracil excision.
Figures



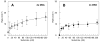

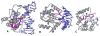
References
-
- David SS, Williams SD. Chemistry of Glycosylases and Endonucleases Involved in Base-Excision Repair. Chem Rev. 1998;98:1221–1262. - PubMed
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutat Res. 2000;460:165–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

