Genome-wide analysis of acetivibrio cellulolyticus provides a blueprint of an elaborate cellulosome system
- PMID: 22646801
- PMCID: PMC3413522
- DOI: 10.1186/1471-2164-13-210
Genome-wide analysis of acetivibrio cellulolyticus provides a blueprint of an elaborate cellulosome system
Abstract
Background: Microbial degradation of plant cell walls and its conversion to sugars and other byproducts is a key step in the carbon cycle on Earth. In order to process heterogeneous plant-derived biomass, specialized anaerobic bacteria use an elaborate multi-enzyme cellulosome complex to synergistically deconstruct cellulosic substrates. The cellulosome was first discovered in the cellulolytic thermophile, Clostridium thermocellum, and much of our knowledge of this intriguing type of protein composite is based on the cellulosome of this environmentally and biotechnologically important bacterium. The recently sequenced genome of the cellulolytic mesophile, Acetivibrio cellulolyticus, allows detailed comparison of the cellulosomes of these two select cellulosome-producing bacteria.
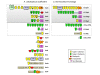
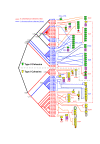
Results: Comprehensive analysis of the A. cellulolyticus draft genome sequence revealed a very sophisticated cellulosome system. Compared to C. thermocellum, the cellulosomal architecture of A. cellulolyticus is much more extensive, whereby the genome encodes for twice the number of cohesin- and dockerin-containing proteins. The A. cellulolyticus genome has thus evolved an inflated number of 143 dockerin-containing genes, coding for multimodular proteins with distinctive catalytic and carbohydrate-binding modules that play critical roles in biomass degradation. Additionally, 41 putative cohesin modules distributed in 16 different scaffoldin proteins were identified in the genome, representing a broader diversity and modularity than those of Clostridium thermocellum. Although many of the A. cellulolyticus scaffoldins appear in unconventional modular combinations, elements of the basic structural scaffoldins are maintained in both species. In addition, both species exhibit similarly elaborate cell-anchoring and cellulosome-related gene- regulatory elements.
Conclusions: This work portrays a particularly intricate, cell-surface cellulosome system in A. cellulolyticus and provides a blueprint for examining the specific roles of the various cellulosomal components in the degradation of complex carbohydrate substrates of the plant cell wall by the bacterium.
Figures

References
-
- Himmel ME, Xu Q, Luo Y, Ding S-Y, Lamed R, Bayer EA. Microbial enzyme systems for biomass conversion: Emerging paradigms. Biofuels. 2010;1:323–341. doi: 10.4155/bfs.09.25. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

