Molecular ecological network analyses
- PMID: 22646978
- PMCID: PMC3428680
- DOI: 10.1186/1471-2105-13-113
Molecular ecological network analyses
Abstract
Background: Understanding the interaction among different species within a community and their responses to environmental changes is a central goal in ecology. However, defining the network structure in a microbial community is very challenging due to their extremely high diversity and as-yet uncultivated status. Although recent advance of metagenomic technologies, such as high throughout sequencing and functional gene arrays, provide revolutionary tools for analyzing microbial community structure, it is still difficult to examine network interactions in a microbial community based on high-throughput metagenomics data.
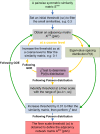
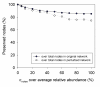
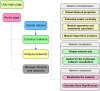
Results: Here, we describe a novel mathematical and bioinformatics framework to construct ecological association networks named molecular ecological networks (MENs) through Random Matrix Theory (RMT)-based methods. Compared to other network construction methods, this approach is remarkable in that the network is automatically defined and robust to noise, thus providing excellent solutions to several common issues associated with high-throughput metagenomics data. We applied it to determine the network structure of microbial communities subjected to long-term experimental warming based on pyrosequencing data of 16 S rRNA genes. We showed that the constructed MENs under both warming and unwarming conditions exhibited topological features of scale free, small world and modularity, which were consistent with previously described molecular ecological networks. Eigengene analysis indicated that the eigengenes represented the module profiles relatively well. In consistency with many other studies, several major environmental traits including temperature and soil pH were found to be important in determining network interactions in the microbial communities examined. To facilitate its application by the scientific community, all these methods and statistical tools have been integrated into a comprehensive Molecular Ecological Network Analysis Pipeline (MENAP), which is open-accessible now (http://ieg2.ou.edu/MENA).
Conclusions: The RMT-based molecular ecological network analysis provides powerful tools to elucidate network interactions in microbial communities and their responses to environmental changes, which are fundamentally important for research in microbial ecology and environmental microbiology.
Figures



References
-
- May RM. Stability and complexity in model ecosystems. Princeton University Press, Princeton, New Jersey; 1973.
-
- Pim S. Food webs. Chapman & Hall, London; 1982.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

