MAPK14/p38α confers irinotecan resistance to TP53-defective cells by inducing survival autophagy
- PMID: 22647487
- PMCID: PMC3429546
- DOI: 10.4161/auto.20268
MAPK14/p38α confers irinotecan resistance to TP53-defective cells by inducing survival autophagy
Abstract
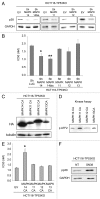
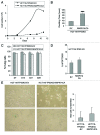
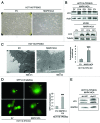
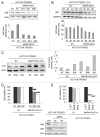
Recently we have shown that the mitogen-activated protein kinase (MAPK) MAPK14/p38α is involved in resistance of colon cancer cells to camptothecin-related drugs. Here we further investigated the cellular mechanisms involved in such drug resistance and showed that, in HCT116 human colorectal adenocarcinoma cells in which TP53 was genetically ablated (HCT116-TP53KO), overexpression of constitutively active MAPK14/p38α decreases cell sensitivity to SN-38 (the active metabolite of irinotecan), inhibits cell proliferation and induces survival-autophagy. Since autophagy is known to facilitate cancer cell resistance to chemotherapy and radiation treatment, we then investigated the relationship between MAPK14/p38α, autophagy and resistance to irinotecan. We demonstrated that induction of autophagy by SN38 is dependent on MAPK14/p38α activation. Finally, we showed that inhibition of MAPK14/p38α or autophagy both sensitizes HCT116-TP53KO cells to drug therapy. Our data proved that the two effects are interrelated, since the role of autophagy in drug resistance required the MAPK14/p38α. Our results highlight the existence of a new mechanism of resistance to camptothecin-related drugs: upon SN38 induction, MAPK14/p38α is activated and triggers survival-promoting autophagy to protect tumor cells against the cytotoxic effects of the drug. Colon cancer cells could thus be sensitized to drug therapy by inhibiting either MAPK14/p38 or autophagy.
Keywords: MAPK14/p38; chemotherapy; colon cancer; irinotecan resistance; survival autophagy.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
