Myotubularin-related protein (MTMR) 9 determines the enzymatic activity, substrate specificity, and role in autophagy of MTMR8
- PMID: 22647598
- PMCID: PMC3386095
- DOI: 10.1073/pnas.1207021109
Myotubularin-related protein (MTMR) 9 determines the enzymatic activity, substrate specificity, and role in autophagy of MTMR8
Abstract
The myotubularins are a large family of inositol polyphosphate 3-phosphatases that, despite having common substrates, subsume unique functions in cells that are disparate. The myotubularin family consists of 16 different proteins, 9 members of which possess catalytic activity, dephosphorylating phosphatidylinositol 3-phosphate [PtdIns(3)P] and phosphatidylinositol 3,5-bisphosphate [PtdIns(3,5)P(2)] at the D-3 position. Seven members are inactive because they lack the conserved cysteine residue in the CX(5)R motif required for activity. We studied a subfamily of homologous myotubularins, including myotubularin-related protein 6 (MTMR6), MTMR7, and MTMR8, all of which dimerize with the catalytically inactive MTMR9. Complex formation between the active myotubularins and MTMR9 increases their catalytic activity and alters their substrate specificity, wherein the MTMR6/R9 complex prefers PtdIns(3,5)P(2) as substrate; the MTMR8/R9 complex prefers PtdIns(3)P. MTMR9 increased the enzymatic activity of MTMR6 toward PtdIns(3,5)P(2) by over 30-fold, and enhanced the activity toward PtdIns(3)P by only 2-fold. In contrast, MTMR9 increased the activity of MTMR8 by 1.4-fold and 4-fold toward PtdIns(3,5)P(2) and PtdIns(3)P, respectively. In cells, the MTMR6/R9 complex significantly increases the cellular levels of PtdIns(5)P, the product of PI(3,5)P(2) dephosphorylation, whereas the MTMR8/R9 complex reduces cellular PtdIns(3)P levels. Consequentially, the MTMR6/R9 complex serves to inhibit stress-induced apoptosis and the MTMR8/R9 complex inhibits autophagy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

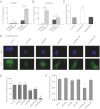

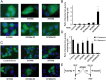
Similar articles
-
MTMR9 increases MTMR6 enzyme activity, stability, and role in apoptosis.J Biol Chem. 2009 Jan 23;284(4):2064-71. doi: 10.1074/jbc.M804292200. Epub 2008 Nov 27. J Biol Chem. 2009. PMID: 19038970 Free PMC article.
-
Condition-dependent functional shift of two Drosophila Mtmr lipid phosphatases in autophagy control.Autophagy. 2021 Dec;17(12):4010-4028. doi: 10.1080/15548627.2021.1899681. Epub 2021 Mar 28. Autophagy. 2021. PMID: 33779490 Free PMC article.
-
Human myotubularin-related protein 9 regulates ER-to-Golgi trafficking and modulates WNT3A secretion.Exp Cell Res. 2020 Jan 1;386(1):111709. doi: 10.1016/j.yexcr.2019.111709. Epub 2019 Nov 6. Exp Cell Res. 2020. PMID: 31704058
-
Myotubularins, a large disease-associated family of cooperating catalytically active and inactive phosphoinositides phosphatases.Hum Mol Genet. 2003 Oct 15;12 Spec No 2:R285-92. doi: 10.1093/hmg/ddg273. Epub 2003 Aug 12. Hum Mol Genet. 2003. PMID: 12925573 Review.
-
Interplay between myotubularins and Ca2+ homeostasis.Biochim Biophys Acta Mol Cell Res. 2024 Jun;1871(5):119739. doi: 10.1016/j.bbamcr.2024.119739. Epub 2024 May 6. Biochim Biophys Acta Mol Cell Res. 2024. PMID: 38710289 Review.
Cited by
-
Loss of catalytically inactive lipid phosphatase myotubularin-related protein 12 impairs myotubularin stability and promotes centronuclear myopathy in zebrafish.PLoS Genet. 2013 Jun;9(6):e1003583. doi: 10.1371/journal.pgen.1003583. Epub 2013 Jun 20. PLoS Genet. 2013. PMID: 23818870 Free PMC article.
-
Phosphatidylinositol 3-phosphates-at the interface between cell signalling and membrane traffic.EMBO J. 2016 Mar 15;35(6):561-79. doi: 10.15252/embj.201593564. Epub 2016 Feb 17. EMBO J. 2016. PMID: 26888746 Free PMC article. Review.
-
The Myotubularin Related Proteins and the Untapped Interaction Potential of Their Disordered C-Terminal Regions.Proteins. 2025 Apr;93(4):831-854. doi: 10.1002/prot.26774. Epub 2024 Nov 30. Proteins. 2025. PMID: 39614773 Free PMC article.
-
Mitochondrial damage triggers the concerted degradation of negative regulators of neuronal autophagy.Nat Commun. 2025 Aug 9;16(1):7367. doi: 10.1038/s41467-025-62379-5. Nat Commun. 2025. PMID: 40783388 Free PMC article.
-
X-linked myotubular myopathy in Rottweiler dogs is caused by a missense mutation in Exon 11 of the MTM1 gene.Skelet Muscle. 2015 Jan 27;5(1):1. doi: 10.1186/s13395-014-0025-3. eCollection 2015. Skelet Muscle. 2015. PMID: 25664165 Free PMC article.
References
-
- Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. - PubMed
-
- Steck PA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. - PubMed
-
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

