Infection of Epstein-Barr virus in a gastric carcinoma cell line induces anchorage independence and global changes in gene expression
- PMID: 22647604
- PMCID: PMC3386136
- DOI: 10.1073/pnas.1202910109
Infection of Epstein-Barr virus in a gastric carcinoma cell line induces anchorage independence and global changes in gene expression
Abstract
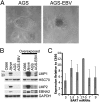
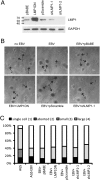
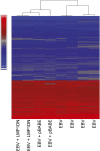
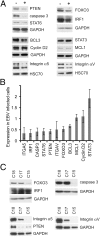
Latent infection of EBV is linked to the development of multiple cancers that have distinct patterns of expression of viral proteins and microRNAs (miRNAs). In this study, we show that in vitro infection of a gastric epithelial cell line with EBV alters growth properties and induces growth in soft agar. The infected cells have high levels of expression of a large cluster of viral miRNAs, [the BamHI A rightward transcript (BART) miRNAs] and limited viral protein expression. Expression profile microarray analysis of this cell line revealed a large number of changes in cellular expression, with decreased expression of many genes. Inhibition of the trace-expressed levels of the viral oncoprotein, latent membrane protein 1, did not affect growth or alter the pattern of cellular expression. The expression changes are highly enriched for genes involved in cell motility and transformation pathways, suggesting these changes are important for the altered growth phenotype. Importantly, the transcripts decreased by microarray are significantly enriched in both experimentally and bioinformatically predicted BART miRNA targets. The absence of viral protein expression and the enrichment for viral miRNA targets in the modulated cell genes suggest that the BART miRNAs are major contributors to the transformed growth properties of the EBV-infected cells. The ability to affect cell growth through miRNA expression without viral protein expression would be a major factor in the development of cancer in individuals with functional immune systems.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Rickinson AB, Kieff E. Epstein-Barr virus. In: Knipe DM, Howley PM, editors. Field’s Virology. 4th Ed. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2575–2627.
-
- Raab-Traub N. Epstein-Barr virus in the pathogenesis of NPC. Semin Cancer Biol. 2002;12:431–441. - PubMed
-
- Lee HS, Chang MS, Yang HK, Lee BL, Kim WH. Epstein-barr virus-positive gastric carcinoma has a distinct protein expression profile in comparison with epstein-barr virus-negative carcinoma. Clin Cancer Res. 2004;10:1698–1705. - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

