Transiently populated intermediate functions as a branching point of the FF domain folding pathway
- PMID: 22647611
- PMCID: PMC3497754
- DOI: 10.1073/pnas.1201799109
Transiently populated intermediate functions as a branching point of the FF domain folding pathway
Abstract
Studies of protein folding and the intermediates that are formed along the folding pathway provide valuable insights into the process by which an unfolded ensemble forms a functional native conformation. However, because intermediates on folding pathways can serve as initiation points of aggregation (implicated in a number of diseases), their characterization assumes an even greater importance. Establishing the role of such intermediates in folding, misfolding, and aggregation remains a major challenge due to their often low populations and short lifetimes. We recently used NMR relaxation dispersion methods and computational techniques to determine an atomic resolution structure of the folding intermediate of a small protein module--the FF domain--with an equilibrium population of 2-3% and a millisecond lifetime, 25 °C. Based on this structure a variant FF domain has been designed in which the native state is selectively destabilized by removing the carboxyl-terminal helix in the native structure to produce a highly populated structural mimic of the intermediate state. Here, we show via solution NMR studies of the designed mimic that the mimic forms distinct conformers corresponding to monomeric and dimeric (K(d) = 0.2 mM) forms of the protein. The conformers exchange on the seconds timescale with a monomer association rate of 1.1 · 10(4) M(-1) s(-1) and with a region responsible for dimerization localized to the amino-terminal residues of the FF domain. This study establishes the FF domain intermediate as a central player in both folding and misfolding pathways and illustrates how incomplete folding can lead to the formation of higher-order structures.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

 HSQC spectrum of the truncated variant, FF1–60, a mimic of the folding intermediate of the full-length domain (11.7 T, 25 °C). Two sets of signals are observed, derived from two separate conformations of the protein, denoted as M and D. (C) Backbone
HSQC spectrum of the truncated variant, FF1–60, a mimic of the folding intermediate of the full-length domain (11.7 T, 25 °C). Two sets of signals are observed, derived from two separate conformations of the protein, denoted as M and D. (C) Backbone  , 15N,
, 15N,  ,
,  , and
, and  chemical shift differences between I and N states of the full-length FF domain (FF1–71) obtained from RD NMR data (13) (
chemical shift differences between I and N states of the full-length FF domain (FF1–71) obtained from RD NMR data (13) ( ), plotted vs. chemical differences between FF1–60 (M form) and the native state FF1–71 (
), plotted vs. chemical differences between FF1–60 (M form) and the native state FF1–71 ( ). ϖstd,i is a nucleus- and residue-specific normalization value that corresponds to the range of shift values (1 SD) that are observed in a database of protein chemical shifts (
). ϖstd,i is a nucleus- and residue-specific normalization value that corresponds to the range of shift values (1 SD) that are observed in a database of protein chemical shifts (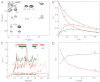
 magnetization exchange spectrum (24, 25) of 0.34 mM FF1–60 recorded with a mixing time T = 0.34 s (11.7 T, 25 °C), showing two sets of autopeaks (MM and DD) connected by exchange cross-peaks (MD and DM) for Ala34. (B) Mixing time, T, dependencies of autopeak (circles) and exchange cross-peak (boxes) volumes for Ala34 from magnetization exchange experiments. The solid lines are generated from a least-squares fit of the exchange data for Ala34 to Eq. S2 in
magnetization exchange spectrum (24, 25) of 0.34 mM FF1–60 recorded with a mixing time T = 0.34 s (11.7 T, 25 °C), showing two sets of autopeaks (MM and DD) connected by exchange cross-peaks (MD and DM) for Ala34. (B) Mixing time, T, dependencies of autopeak (circles) and exchange cross-peak (boxes) volumes for Ala34 from magnetization exchange experiments. The solid lines are generated from a least-squares fit of the exchange data for Ala34 to Eq. S2 in  correlation spectra measured in conventional 15N R1 and R1ρ experiments (28, 29). As discussed in the text, the apparent 15N R1 and R2 rates are affected by slow exchange between M and D states; intrinsic relaxation rates (solid lines) are calculated using the procedure described in
correlation spectra measured in conventional 15N R1 and R1ρ experiments (28, 29). As discussed in the text, the apparent 15N R1 and R2 rates are affected by slow exchange between M and D states; intrinsic relaxation rates (solid lines) are calculated using the procedure described in  HSQC spectra of FF1–60 recorded as a function of protein concentration. The volumes are normalized by total protein concentration.
HSQC spectra of FF1–60 recorded as a function of protein concentration. The volumes are normalized by total protein concentration.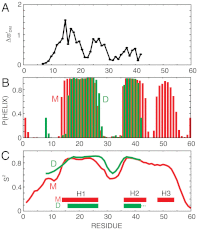
 , as described in the text. Values of
, as described in the text. Values of  are shown for residues Thr8-Met42; the chemical shifts of the first seven residues are indistinguishable in the two forms, while those from the C terminus (residues Ile43-Gln60) are missing in NMR spectra of the dimer. (B) TALOS-plus predicted (33) α-helix probability, P (helix), plotted vs. residue for states M (red) and D (green, residues Thr8-Met42 only). (C) RCI-predicted (34, 35) order parameters, S2, for the backbone amide groups of M (red) and D (green).
are shown for residues Thr8-Met42; the chemical shifts of the first seven residues are indistinguishable in the two forms, while those from the C terminus (residues Ile43-Gln60) are missing in NMR spectra of the dimer. (B) TALOS-plus predicted (33) α-helix probability, P (helix), plotted vs. residue for states M (red) and D (green, residues Thr8-Met42 only). (C) RCI-predicted (34, 35) order parameters, S2, for the backbone amide groups of M (red) and D (green).References
-
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. - PubMed
-
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. - PubMed
-
- Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5:15–22. - PubMed
-
- Korzhnev DM, Kay LE. Probing invisible, low-populated states of protein molecules by relaxation dispersion NMR spectroscopy: An application to protein folding. Acc Chem Res. 2008;41:442–451. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

