Product binding varies dramatically between processive and nonprocessive cellulase enzymes
- PMID: 22648408
- PMCID: PMC3397907
- DOI: 10.1074/jbc.M112.365510
Product binding varies dramatically between processive and nonprocessive cellulase enzymes
Abstract
Cellulases hydrolyze β-1,4 glycosidic linkages in cellulose, which are among the most prevalent and stable bonds in Nature. Cellulases comprise many glycoside hydrolase families and exist as processive or nonprocessive enzymes. Product inhibition negatively impacts cellulase action, but experimental measurements of product-binding constants vary significantly, and there is little consensus on the importance of this phenomenon. To provide molecular level insights into cellulase product inhibition, we examine the impact of product binding on processive and nonprocessive cellulases by calculating the binding free energy of cellobiose to the product sites of catalytic domains of processive and nonprocessive enzymes from glycoside hydrolase families 6 and 7. The results suggest that cellobiose binds to processive cellulases much more strongly than nonprocessive cellulases. We also predict that the presence of a cellodextrin bound in the reactant site of the catalytic domain, which is present during enzymatic catalysis, has no effect on product binding in nonprocessive cellulases, whereas it significantly increases product binding to processive cellulases. This difference in product binding correlates with hydrogen bonding between the substrate-side ligand and the cellobiose product in processive cellulase tunnels and the additional stabilization from the longer tunnel-forming loops. The hydrogen bonds between the substrate- and product-side ligands are disrupted by water in nonprocessive cellulase clefts, and the lack of long tunnel-forming loops results in lower affinity of the product ligand. These findings provide new insights into the large discrepancies reported for binding constants for cellulases and suggest that product inhibition will vary significantly based on the amount of productive binding for processive cellulases on cellulose.
Figures

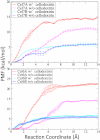

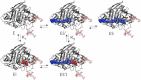
References
-
- Himmel M. E., Ding S. Y., Johnson D. K., Adney W. S., Nimlos M. R., Brady J. W., Foust T. D. (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315, 804–807 - PubMed
-
- Andrić P., Meyer A. S., Jensen P. A., Dam-Johansen K. (2010) Reactor design for minimizing product inhibition during enzymatic lignocellulose hydrolysis. I. Significance and mechanism of cellobiose and glucose inhibition on cellulolytic enzymes. Biotechnol. Adv. 28, 308–324 - PubMed
-
- Bansal P., Hall M., Realff M. J., Lee J. H., Bommarius A. S. (2009) Modeling cellulase kinetics on lignocellulosic substrates. Biotechnol. Adv. 27, 833–848 - PubMed
-
- Gan Q., Allen S. J., Taylor G. (2003) Kinetic dynamics in heterogeneous enzymatic hydrolysis of cellulose: an overview, an experimental study and mathematical modelling. Process Biochemistry 38, 1003–1018
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

