Comparing enhancer action in cis and in trans
- PMID: 22649083
- PMCID: PMC3415998
- DOI: 10.1534/genetics.112.140954
Comparing enhancer action in cis and in trans
Abstract
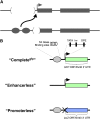
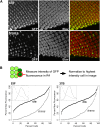
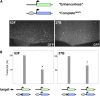
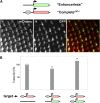
Studies from diverse systems have shown that distinct interchromosomal interactions are a central component of nuclear organization. In some cases, these interactions allow an enhancer to act in trans, modulating the expression of a gene encoded on a separate chromosome held in close proximity. Despite recent advances in uncovering such phenomena, our understanding of how a regulatory element acts on another chromosome remains incomplete. Here, we describe a transgenic approach to better understand enhancer action in trans in Drosophila melanogaster. Using phiC31-based recombinase-mediated cassette exchange (RMCE), we placed transgenes carrying combinations of the simple enhancer GMR, a minimal promoter, and different fluorescent reporters at equivalent positions on homologous chromosomes so that they would pair via the endogenous somatic pairing machinery of Drosophila. Our data demonstrate that the enhancer GMR is capable of activating a promoter in trans and does so in a variegated pattern, suggesting stochastic interactions between the enhancer and the promoter when they are carried on separate chromosomes. Furthermore, we quantitatively assessed the impact of two concurrent promoter targets in cis and in trans to GMR, demonstrating that each promoter is capable of competing for the enhancer's activity, with the presence of one negatively affecting expression from the other. Finally, the single-cell resolution afforded by our approach allowed us to show that promoters in cis and in trans to GMR can both be activated in the same nucleus, implying that a single enhancer can share its activity between multiple promoter targets carried on separate chromosomes.
Figures


Comment in
-
Transvection in 2012: site-specific transgenes reveal a plethora of trans-regulatory effects.Genetics. 2012 Aug;191(4):1037-9. doi: 10.1534/genetics.112.142893. Genetics. 2012. PMID: 22879406 Free PMC article. No abstract available.
References
-
- Apostolou E., Thanos D., 2008. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell 134: 85–96 - PubMed
-
- Bacher C. P., Guggiari M., Brors B., Augui S., Clerc P., et al. , 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8: 293–299 - PubMed
-
- Bantignies F., Cavalli G., 2011. Polycomb group proteins: repression in 3D. Trends Genet. 27: 454–464 - PubMed
-
- Bartkuhn M., Renkawitz R., 2008. Long range chromatin interactions involved in gene regulation. Biochim. Biophys. Acta 1783: 2161–2166 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

