Single-molecule Förster resonance energy transfer reveals an innate fidelity checkpoint in DNA polymerase I
- PMID: 22650319
- PMCID: PMC3448555
- DOI: 10.1021/ja3038273
Single-molecule Förster resonance energy transfer reveals an innate fidelity checkpoint in DNA polymerase I
Abstract
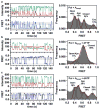
Enzymatic reactions typically involve complex dynamics during substrate binding, conformational rearrangement, chemistry, and product release. The noncovalent steps provide kinetic checkpoints that contribute to the overall specificity of enzymatic reactions. DNA polymerases perform DNA replication with outstanding fidelity by actively rejecting noncognate nucleotide substrates early in the reaction pathway. Substrates are delivered to the active site by a flexible fingers subdomain of the enzyme, as it converts from an open to a closed conformation. The conformational dynamics of the fingers subdomain might also play a role in nucleotide selection, although the precise role is currently unknown. Using single-molecule Förster resonance energy transfer, we observed individual Escherichia coli DNA polymerase I (Klenow fragment) molecules performing substrate selection. We discovered that the fingers subdomain actually samples through three distinct conformations--open, closed, and a previously unrecognized intermediate conformation. We measured the overall dissociation rate of the polymerase-DNA complex and the distribution among the various conformational states in the absence and presence of nucleotide substrates, which were either correct or incorrect. Correct substrates promote rapid progression of the polymerase to the catalytically competent closed conformation, whereas incorrect nucleotides block the enzyme in the intermediate conformation and induce rapid dissociation from DNA. Remarkably, incorrect nucleotide substrates also promote partitioning of DNA to the spatially separated 3'-5' exonuclease domain, providing an additional mechanism to prevent misincorporation at the polymerase active site. These results reveal the existence of an early innate fidelity checkpoint, rejecting incorrect nucleotide substrates before the enzyme encloses the nascent base pair.
Figures
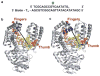


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

