Autophagy is active in normal colon mucosa
- PMID: 22652752
- PMCID: PMC3427255
- DOI: 10.4161/auto.19738
Autophagy is active in normal colon mucosa
Abstract
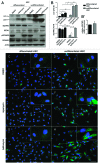
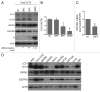
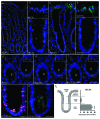
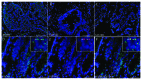
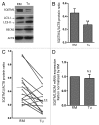
Recently, autophagy has been found to be strongly activated in colon cancer cells, but few studies have addressed the normal colon mucosa. The aim of this study was to characterize autophagy in normal human intestinal cells. We used the expression of LC3-II and BECN1 as well as SQSTM1 as markers of autophagy activity. Using the normal human intestinal epithelial crypt (HIEC) cell experimental model, we found that autophagy was much more active in undifferentiated cells than in differentiated cells. In the normal adult colonic mucosa, BECN1 was found in the proliferative epithelial cells of the lower part of the gland while SQSTM1 was predominantly found in the differentiated cells of the upper part of the gland and surface epithelium. Interestingly, the weak punctate pattern of SQSTM1 expression in the lower gland colocalized with BECN1-labeled autophagosomes. The usefulness of SQSTM1 as an active autophagy marker was confirmed in colon cancer specimens at the protein and transcript levels. In conclusion, our results show that autophagy is active in the colonic gland and is associated with the intestinal proliferative/undifferentiated and progenitor cell populations.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
