The Fbw7 and betaTRCP E3 ubiquitin ligases and their roles in tumorigenesis
- PMID: 22652772
- PMCID: PMC3374336
- DOI: 10.2741/4045
The Fbw7 and betaTRCP E3 ubiquitin ligases and their roles in tumorigenesis
Abstract
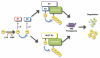
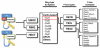
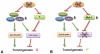
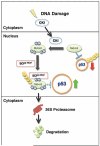
The Ubiquitin Proteasome System (UPS) is a major regulator of protein abundance in the cell. The UPS influences the functions of multiple biological processes by targeting key regulators for destruction. E3 ubiquitin ligases are a vital component of the UPS machinery, working with E1 and E2 enzymes to bind substrates and facilitate the transfer of ubiquitin molecules onto the target protein. This poly-ubiquitination, in turn, directs the modified proteins for proteolysis by the 26S proteasome. As the UPS regulates the degradation of multiple oncogenes and tumor suppressors, the dysregulation of this pathway is known to promote various diseases including cancer. While E1 and E2 enzymes have only been minimally linked to cancer development, burgeoning amounts of evidence have implicated loss or gain of E3 function as a key factor in cancer initiation and progression. This review will examine the literature on two SCF-type E3 ligases, SCFFbw7 and SCFbeta-TRCP. In particular, we will highlight novel substrates recently identified for these two E3 ligases, and further discuss how UPS regulation of these targets may promote carcinogenesis.
Figures

References
-
- Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–53. - PubMed
-
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–39. - PubMed
-
- Nakayama KI, Nakayama K. Regulation of the cell cycle by SCF-type ubiquitin ligases. Semin Cell Dev Biol. 2005;16:323–33. - PubMed
-
- Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

