Comparative genomics reveals two novel RNAi factors in Trypanosoma brucei and provides insight into the core machinery
- PMID: 22654659
- PMCID: PMC3359990
- DOI: 10.1371/journal.ppat.1002678
Comparative genomics reveals two novel RNAi factors in Trypanosoma brucei and provides insight into the core machinery
Abstract
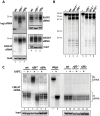
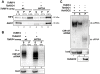
The introduction ten years ago of RNA interference (RNAi) as a tool for molecular exploration in Trypanosoma brucei has led to a surge in our understanding of the pathogenesis and biology of this human parasite. In particular, a genome-wide RNAi screen has recently been combined with next-generation Illumina sequencing to expose catalogues of genes associated with loss of fitness in distinct developmental stages. At present, this technology is restricted to RNAi-positive protozoan parasites, which excludes T. cruzi, Leishmania major, and Plasmodium falciparum. Therefore, elucidating the mechanism of RNAi and identifying the essential components of the pathway is fundamental for improving RNAi efficiency in T. brucei and for transferring the RNAi tool to RNAi-deficient pathogens. Here we used comparative genomics of RNAi-positive and -negative trypanosomatid protozoans to identify the repertoire of factors in T. brucei. In addition to the previously characterized Argonaute 1 (AGO1) protein and the cytoplasmic and nuclear Dicers, TbDCL1 and TbDCL2, respectively, we identified the RNA Interference Factors 4 and 5 (TbRIF4 and TbRIF5). TbRIF4 is a 3'-5' exonuclease of the DnaQ superfamily and plays a critical role in the conversion of duplex siRNAs to the single-stranded form, thus generating a TbAGO1-siRNA complex required for target-specific cleavage. TbRIF5 is essential for cytoplasmic RNAi and appears to act as a TbDCL1 cofactor. The availability of the core RNAi machinery in T. brucei provides a platform to gain mechanistic insights in this ancient eukaryote and to identify the minimal set of components required to reconstitute RNAi in RNAi-deficient parasites.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



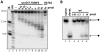
References
-
- DaRocha WD, Otsu K, Teixeira SM, Donelson JE. Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi. Mol Biochem Parasitol. 2004;133:175–186. - PubMed
-
- Robinson KA, Beverley SM. Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania. Mol Biochem Parasitol. 2003;128:217–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

