Early Diagnosis of Werner's Syndrome Using Exome-Wide Sequencing in a Single, Atypical Patient
- PMID: 22654791
- PMCID: PMC3356119
- DOI: 10.3389/fendo.2011.00008
Early Diagnosis of Werner's Syndrome Using Exome-Wide Sequencing in a Single, Atypical Patient
Abstract
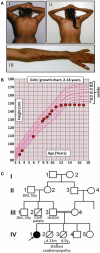
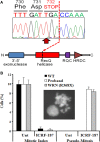
Genetic diagnosis of inherited metabolic disease is conventionally achieved through syndrome recognition and targeted gene sequencing, but many patients receive no specific diagnosis. Next-generation sequencing allied to capture of expressed sequences from genomic DNA now offers a powerful new diagnostic approach. Barriers to routine diagnostic use include cost, and the complexity of interpreting results arising from simultaneous identification of large numbers of variants. We applied exome-wide sequencing to an individual, 16-year-old daughter of consanguineous parents with a novel syndrome of short stature, severe insulin resistance, ptosis, and microcephaly. Pulldown of expressed sequences from genomic DNA followed by massively parallel sequencing was undertaken. Single nucleotide variants were called using SAMtools prior to filtering based on sequence quality and existence in control genomes and exomes. Of 485 genetic variants predicted to alter protein sequence and absent from control data, 24 were homozygous in the patient. One mutation - the p.Arg732X mutation in the WRN gene - has previously been reported in Werner's syndrome (WS). On re-evaluation of the patient several early features of WS were detected including loss of fat from the extremities and frontal hair thinning. Lymphoblastoid cells from the proband exhibited a defective decatenation checkpoint, consistent with loss of WRN activity. We have thus diagnosed WS some 15 years earlier than average, permitting aggressive prophylactic therapy and screening for WS complications, illustrating the potential of exome-wide sequencing to achieve early diagnosis and change management of rare autosomal recessive disease, even in individual patients of consanguineous parentage with apparently novel syndromes.
Keywords: WRN; Werner's syndrome; diabetes; insulin resistance; whole exome sequencing.
Figures


References
-
- Alderton G. K., Galbiati L., Griffith E., Surinya K. H., Neitzel H., Jackson A. P., Jeggo P. A., O'Driscoll M. (2006). Regulation of mitotic entry by microcephalin and its overlap with ATR signalling. Nat. Cell Biol. 8, 725–733 - PubMed
-
- Bilgüvar K., Oztürk A. K., Louvi A., Kwan K. Y., Choi M., Tatli B., Yalnizoğlu D., Tüysüz B., Cağlayan A. O., Gökben S., Kaymakçalan H., Barak T., Bakircioğlu M., Yasuno K., Ho W., Sanders S., Zhu Y., Yilmaz S., Dinçer A., Johnson M. H., Bronen R. A., Koçer N., Per H., Mane S., Pamir M. N., Yalçinkaya C., Kumandaş S., Topçu M., Ozmen M., Sestan N., Lifton R. P., State M. W., Günel M. (2010). Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467, 207–210 10.1038/nature09327 - DOI - PMC - PubMed
-
- Bonnefond A., Durand E., Sand O., Graeve F. D., Gallina S., Busiah K., Lobbens S., Simon A., Bellanné-Chantelot C., Létourneau L., Scharfmann R., Delplanque J., Sladek R., Polak M., Vaxillaire M., Froguel P. (2010). Molecular diagnosis of neonatal diabetes mellitus using next-generation sequencing of the whole exome. PLoS ONE 5, e13630. 10.1371/journal.pone.0013630 - DOI - PMC - PubMed
-
- Choi M., Scholl U. I., Ji W., Liu T., Tikhonova I. R., Zumbo P., Nayir A., Bakkaloğlu A. i., Ozen S., Sanjad S., Nelson-Williams C., Farhi A., Mane S., Lifton R. P. (2009). Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 106, 19096–19101 - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

