Insulin-like growth factor binding proteins: a structural perspective
- PMID: 22654863
- PMCID: PMC3356058
- DOI: 10.3389/fendo.2012.00038
Insulin-like growth factor binding proteins: a structural perspective
Abstract
Insulin-like growth factor binding proteins (IGFBP-1 to -6) bind insulin-like growth factors-I and -II (IGF-I and IGF-II) with high affinity. These binding proteins maintain IGFs in the circulation and direct them to target tissues, where they promote cell growth, proliferation, differentiation, and survival via the type 1 IGF receptor. IGFBPs also interact with many other molecules, which not only influence their modulation of IGF action but also mediate IGF-independent activities that regulate processes such as cell migration and apoptosis by modulating gene transcription. IGFBPs-1 to -6 are structurally similar proteins consisting of three distinct domains, N-terminal, linker, and C-terminal. There have been major advances in our understanding of IGFBP structure in the last decade and a half. While there is still no structure of an intact IGFBP, several structures of individual N- and C-domains have been solved. The structure of a complex of N-BP-4:IGF-I:C-BP-4 has also been solved, providing a detailed picture of the structural features of the IGF binding site and the mechanism of binding. Structural studies have also identified features important for interaction with extracellular matrix components and integrins. This review summarizes structural studies reported so far and highlights features important for binding not only IGF but also other partners. We also highlight future directions in which structural studies will add to our knowledge of the role played by the IGFBP family in normal growth and development, as well as in disease.
Keywords: IGF binding protein; insulin-like growth factor; protein structure.
Figures
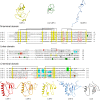
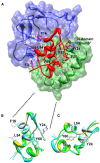
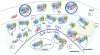
References
-
- Arai T., Clarke J., Parker A., Busby W., Jr., Nam T., Clemmons D. R. (1996b). Substitution of specific amino acids in insulin-like growth factor (IGF) binding protein 5 alters heparin binding and its change in affinity for IGF-I response to heparin. J. Biol. Chem. 271, 6099–6106 10.1074/jbc.271.11.6099 - DOI - PubMed
-
- Arai T., Parker A., Busby W., Jr., Clemmons D. R. (1994). Heparin, heparan sulfate, and dermatan sulfate regulate formation of the insulin-like growth factor-I and insulin-like growth factor-binding protein complexes. J. Biol. Chem. 269, 20388–20393 - PubMed
LinkOut - more resources
Full Text Sources

