Evolution and expression of tandem duplicated maize flavonol synthase genes
- PMID: 22654889
- PMCID: PMC3360202
- DOI: 10.3389/fpls.2012.00101
Evolution and expression of tandem duplicated maize flavonol synthase genes
Abstract
Flavonoids are specialized compounds widely distributed and with diverse functions throughout the plant kingdom and with several benefits for human health. In particular, flavonols, synthesized by flavonol synthase (FLS), protect plants against UV-B radiation and are essential for male fertility in maize and other plants. We have recently characterized a UV-B inducible ZmFLS1, corresponding to the first to be described in monocot plants. Interestingly, the new assembly of the B73 maize genome revealed the presence of a second putative FLS gene (ZmFLS2), with very high identity with ZmFLS1. ZmFLSs expression was analyzed in different maize tissues, and by combining electrophoretic mobility shift assays and transient expression experiments, we show that both genes are direct targets of anthocyanin (C1/PL1 + R/B) and 3-deoxy flavonoid (P1) transcriptional regulators. ZmFLS expression analyses show higher levels of both transcripts in high altitude landraces than inbred lines, and both genes are regulated by UV-B radiation in all lines analyzed. Moreover, the high sequence conservation of the ZmFLS promoters between maize lines suggests that the differences observed in ZmFLS expression are due to allelic variations in the transcription factors that regulate their activities. Finally, we generated pFLS1::FLS1-RFP transgenic plants and analyzed ZmFLS1 expression in different maize tissues; we found that this enzyme is localized in the ER and the perinuclear region.
Keywords: UV-B; duplication; grasses; maize; natural variation.
Figures

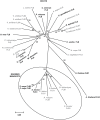
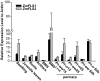
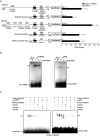

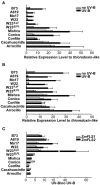










References
-
- Bruggmann R., Bharti A. K., Gundlach H., Lai J., Young S., Pontaroli A. C., Wei F., Haberer G., Fuks G., Du C., Raymond C., Estep M. C., Liu R., Bennetzen J. L., Chan A. P., Rabinowicz P. D., Quackenbush J., Barbazuk W. B., Wing R. A., Birren B., Nusbaum C., Rounsley S., Mayer K. F., Messing J. (2006). Uneven chromosome contraction and expansion in the maize genome. Genome Res. 16, 1241–1251 10.1101/gr.5338906 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

