IMGT-ONTOLOGY 2012
- PMID: 22654892
- PMCID: PMC3358611
- DOI: 10.3389/fgene.2012.00079
IMGT-ONTOLOGY 2012
Abstract
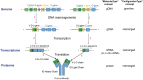
Immunogenetics is the science that studies the genetics of the immune system and immune responses. Owing to the complexity and diversity of the immune repertoire, immunogenetics represents one of the greatest challenges for data interpretation: a large biological expertise, a considerable effort of standardization and the elaboration of an efficient system for the management of the related knowledge were required. IMGT®, the international ImMunoGeneTics information system® (http://www.imgt.org) has reached that goal through the building of a unique ontology, IMGT-ONTOLOGY, which represents the first ontology for the formal representation of knowledge in immunogenetics and immunoinformatics. IMGT-ONTOLOGY manages the immunogenetics knowledge through diverse facets that rely on the seven axioms of the Formal IMGT-ONTOLOGY or IMGT-Kaleidoscope: "IDENTIFICATION," "DESCRIPTION," "CLASSIFICATION," "NUMEROTATION," "LOCALIZATION," "ORIENTATION," and "OBTENTION." The concepts of identification, description, classification, and numerotation generated from the axioms led to the elaboration of the IMGT(®) standards that constitute the IMGT Scientific chart: IMGT®standardized keywords (concepts of identification), IMGT® standardized labels (concepts of description), IMGT® standardized gene and allele nomenclature (concepts of classification) and IMGT unique numbering and IMGT Collier de Perles (concepts of numerotation). IMGT-ONTOLOGY has become the global reference in immunogenetics and immunoinformatics for the knowledge representation of immunoglobulins (IG) or antibodies, T cell receptors (TR), and major histocompatibility (MH) proteins of humans and other vertebrates, proteins of the immunoglobulin superfamily (IgSF) and MH superfamily (MhSF), related proteins of the immune system (RPI) of vertebrates and invertebrates, therapeutic monoclonal antibodies (mAbs), fusion proteins for immune applications (FPIA), and composite proteins for clinical applications (CPCA).
Keywords: IMGT; IMGT-ONTOLOGY; T cell receptor; antibody; immune repertoire; immunogenetics; immunoglobulin; immunoinformatics.
Figures






References
-
- Alamyar E., Giudicelli V., Shuo L., Duroux P., Lefranc M.-P. (2012). IMGT/HighV-QUEST: the IMGT® web portal for immunoglobulin (IG) or antibody and T cell receptor (TR) analysis from NGS high throughput and deep sequencing. Immunome Res. 8, 1–2
-
- Bleakley K., Giudicelli V., Wu Y., Lefranc M.-P., Biau G. (2006). IMGT standardization for statistical analyses of T cell receptor junctions: the TRAV-TRAJ example. In silico Biol. (Gedrukt) 6, 573–588 - PubMed
-
- Bradford Y., Conlin T., Dunn N., Fashena D., Frazer K., Howe D. G., Knight J., Mani P., Martin R., Moxon S. A. T., Paddock H., Pich C., Ramachandran S., Ruef B. J., Ruzicka L., Bauer Schaper H., Schaper K., Shao X., Singer A., Sprague J., Sprunger B., Van Slyke C., Westerfield M. (2011). ZFIN: enhancements and updates to the zebrafish model organism database. Nucleic Acids Res. 39, D822–D829 10.1093/nar/gkq1077 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

