Comparison of storage conditions for human vaginal microbiome studies
- PMID: 22655031
- PMCID: PMC3360033
- DOI: 10.1371/journal.pone.0036934
Comparison of storage conditions for human vaginal microbiome studies
Abstract
Background: The effect of storage conditions on the microbiome and metabolite composition of human biological samples has not been thoroughly investigated as a potential source of bias. We evaluated the effect of two common storage conditions used in clinical trials on the bacterial and metabolite composition of the vaginal microbiota using pyrosequencing of barcoded 16S rRNA gene sequencing and (1)H-NMR analyses.
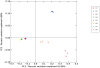
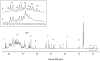
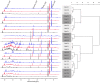
Methodology/principal findings: Eight women were enrolled and four mid-vaginal swabs were collected by a physician from each woman. The samples were either processed immediately, stored at -80°C for 4 weeks or at -20°C for 1 week followed by transfer to -80°C for another 4 weeks prior to analysis. Statistical methods, including Kolmogorovo-Smirnov and Wilcoxon tests, were performed to evaluate the differences in vaginal bacterial community composition and metabolites between samples stored under different conditions. The results showed that there were no significant differences between samples processed immediately after collection or stored for varying durations. (1)H-NMR analysis of the small molecule metabolites in vaginal secretions indicated that high levels of lactic acid were associated with Lactobacillus-dominated communities. Relative abundance of lactic acid did not appear to correlate with relative abundance of individual Lactobacillus sp. in this limited sample, although lower levels of lactic acid were observed when L. gasseri was dominant, indicating differences in metabolic output of seemingly similar communities.
Conclusions/significance: These findings benefit large-scale, field-based microbiome and metabolomic studies of the vaginal microbiota.
Conflict of interest statement
Figures

References
-
- Dolfing J, Vos A, Bloem J, Ehlert PA, Naumova NB, et al. Microbial diversity in archived soils. Science. 2004;306:813. - PubMed
-
- Klammer S, Mondini C, Insam H. Microbial community fingerprints of composts stored under different conditions. Ann Microbiol. 2005;55:299–305.
-
- Nechvatal JM, Ram JL, Basson MD, Namprachan P, Niec SR, et al. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J Microbiol Methods. 2008;72:124–132. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

