ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing
- PMID: 22658618
- PMCID: PMC3391416
- DOI: 10.1016/j.jmoldx.2012.03.002
ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing
Abstract
Lynch syndrome (hereditary nonpolyposis colon cancer) and adenomatous polyposis syndromes frequently have overlapping clinical features. Current approaches for molecular genetic testing are often stepwise, taking a best-candidate gene approach with testing of additional genes if initial results are negative. We report a comprehensive assay called ColoSeq that detects all classes of mutations in Lynch and polyposis syndrome genes using targeted capture and massively parallel next-generation sequencing on the Illumina HiSeq2000 instrument. In blinded specimens and colon cancer cell lines with defined mutations, ColoSeq correctly identified 28/28 (100%) pathogenic mutations in MLH1, MSH2, MSH6, PMS2, EPCAM, APC, and MUTYH, including single nucleotide variants (SNVs), small insertions and deletions, and large copy number variants. There was 100% reproducibility of detection mutation between independent runs. The assay correctly identified 222 of 224 heterozygous SNVs (99.4%) in HapMap samples, demonstrating high sensitivity of calling all variants across each captured gene. Average coverage was greater than 320 reads per base pair when the maximum of 96 index samples with barcodes were pooled. In a specificity study of 19 control patients without cancer from different ethnic backgrounds, we did not find any pathogenic mutations but detected two variants of uncertain significance. ColoSeq offers a powerful, cost-effective means of genetic testing for Lynch and polyposis syndromes that eliminates the need for stepwise testing and multiple follow-up clinical visits.
Copyright © 2012 American Society for Investigative Pathology and the Association for Molecular Pathology. Published by Elsevier Inc. All rights reserved.
Figures
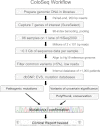



References
-
- Jenkins M.A., Hayashi S., O'Shea A.M., Burgart L.J., Smyrk T.C., Shimizu D., Waring P.M., Ruszkiewicz A.R., Pollett A.F., Redston M., Barker M.A., Baron J.A., Casey G.R., Dowty J.G., Giles G.G., Limburg P., Newcomb P., Young J.P., Walsh M.D., Thibodeau S.N., Lindor N.M., Lemarchand L., Gallinger S., Haile R.W., Potter J.D., Hopper J.L., Jass J.R. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population-based study. Gastroenterology. 2007;133:48–56. - PMC - PubMed
-
- Kohlmann W., Gruber S.B. Copyright University of Washington; Seattle: 1993–2012. Lynch Syndrome: In GeneReviews [Internet]http://www.ncbi.nlm.nih.gov/books/NBK1211 Last revised August 11, 2011.
-
- American Gastroenterological Association medical position statement: hereditary colorectal cancer and genetic testing. Gastroenterology. 2001;121:195–197. - PubMed
-
- Cao Y., Pieretti M., Marshall J., Khattar N.H., Chen B., Kam-Morgan L., Lynch H. Challenge in the differentiation between attenuated familial adenomatous polyposis and hereditary nonpolyposis colorectal cancer: case report with review of the literature. Am J Gastroenterol. 2002;97:1822–1827. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

