Recent developments in single-molecule DNA mechanics
- PMID: 22658779
- PMCID: PMC3726534
- DOI: 10.1016/j.sbi.2012.04.007
Recent developments in single-molecule DNA mechanics
Abstract
Over the past two decades, measurements on individual stretched and twisted DNA molecules have helped define the basic elastic properties of the double helix and enabled real-time functional assays of DNA-associated molecular machines. Recently, new magnetic tweezers approaches for simultaneously measuring freely fluctuating twist and extension have begun to shed light on the structural dynamics of large nucleoprotein complexes. Related technical advances have facilitated direct measurements of DNA torque, contributing to a better understanding of abrupt structural transitions in mechanically stressed DNA. The new measurements have also been exploited in studies that hint at a developing synergistic relationship between single-molecule manipulation and structural DNA nanotechnology.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


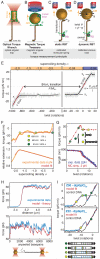
 (where l is the length of the DNA and Pt,eff is the effective twist persistence length). Δθ0,L, extrapolated change in twist per basepair for the B-L transition; N, number of basepairs of DNA; κL, torsional spring constant for L-DNA. Data are from [26], obtained using dynamic RBT. F) Torque and extension for a complete B-to-L transition measured using an optical torque wrench, and global fit to an analytical model[34]. G) Torque and extension at the plectonemic buckling transition. Extension data are shown together with simulated extension and simulated torque[30]. H) Sequence dependent unpeeling of dsDNA under extension[40], shown together with an equilibrium model based on known basepair stabilities. I) The torsional response of GC repeats[26] is explained by cooperative B-Z transitions. In the depicted microstates, energetically costly junctions between B-form and Z-form are shown as yellow bars.
(where l is the length of the DNA and Pt,eff is the effective twist persistence length). Δθ0,L, extrapolated change in twist per basepair for the B-L transition; N, number of basepairs of DNA; κL, torsional spring constant for L-DNA. Data are from [26], obtained using dynamic RBT. F) Torque and extension for a complete B-to-L transition measured using an optical torque wrench, and global fit to an analytical model[34]. G) Torque and extension at the plectonemic buckling transition. Extension data are shown together with simulated extension and simulated torque[30]. H) Sequence dependent unpeeling of dsDNA under extension[40], shown together with an equilibrium model based on known basepair stabilities. I) The torsional response of GC repeats[26] is explained by cooperative B-Z transitions. In the depicted microstates, energetically costly junctions between B-form and Z-form are shown as yellow bars.
References
-
- Bustamante C, Bryant Z, Smith SB. Ten years of tension: Single-molecule DNA mechanics. Nature. 2003;421(6921):423–427. - PubMed
-
-
Vijayan V, Zuzow R, O’Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc Natl Acad Sci U S A. 2009;106(52):22564–22568. A dramatic demonstration of an organism using mechanical stress in DNA to control complex global changes in transcriptional profiles. The physical basis for differing promoter responses, which are only hinted at by observed sequence biases, deserves further study.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

