Fluorescence polarization assay for inhibitors of the kinase domain of receptor interacting protein 1
- PMID: 22658960
- PMCID: PMC3398245
- DOI: 10.1016/j.ab.2012.05.019
Fluorescence polarization assay for inhibitors of the kinase domain of receptor interacting protein 1
Abstract
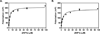
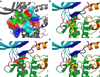
Necrotic cell death is prevalent in many different pathological disease states and in traumatic injury. Necroptosis is a form of necrosis that stems from specific signaling pathways, with the key regulator being receptor interacting protein 1 (RIP1), a serine/threonine kinase. Specific inhibitors of RIP1, termed necrostatins, are potent inhibitors of necroptosis. Necrostatins are structurally distinct from one another yet still possess the ability to inhibit RIP1 kinase activity. To further understand the differences in the binding of the various necrostatins to RIP1 and to develop a robust high-throughput screening (HTS) assay, which can be used to identify new classes of RIP1 inhibitors, we synthesized fluorescein derivatives of Necrostatin-1 (Nec-1) and Nec-3. These compounds were used to establish a fluorescence polarization (FP) assay to directly measure the binding of necrostatins to RIP1 kinase. The fluorescein-labeled compounds are well suited for HTS because the assays have a dimethyl sulfoxide (DMSO) tolerance up to 5% and Z' scores of 0.62 (fluorescein-Nec-1) and 0.57 (fluorescein-Nec-3). In addition, results obtained from the FP assays and ligand docking studies provide insights into the putative binding sites of Nec-1, Nec-3, and Nec-4.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures





References
-
- Dunai Z, Bauer PI, Mihalik R. Necroptosis: Biochemical, Physiological and Pathological Aspects. Pathol. Oncol. Res. 2011 - PubMed
-
- Galluzzi L, Vanden Berghe T, Vanlangenakker N, Buettner S, Eisenberg T, Vandenabeele P, Madeo F, Kroemer G. Programmed necrosis from molecules to health and disease. Int. Rev. Cell Mol. Biol. 2011;289:1–35. - PubMed
-
- Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. - PubMed
-
- He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

