Single-cell systems biology by super-resolution imaging and combinatorial labeling
- PMID: 22660740
- PMCID: PMC3418883
- DOI: 10.1038/nmeth.2069
Single-cell systems biology by super-resolution imaging and combinatorial labeling
Abstract
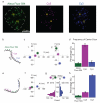
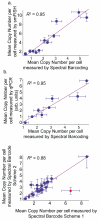
Fluorescence microscopy is a powerful quantitative tool for exploring regulatory networks in single cells. However, the number of molecular species that can be measured simultaneously is limited by the spectral overlap between fluorophores. Here we demonstrate a simple but general strategy to drastically increase the capacity for multiplex detection of molecules in single cells by using optical super-resolution microscopy (SRM) and combinatorial labeling. As a proof of principle, we labeled mRNAs with unique combinations of fluorophores using fluorescence in situ hybridization (FISH), and resolved the sequences and combinations of fluorophores with SRM. We measured mRNA levels of 32 genes simultaneously in single Saccharomyces cerevisiae cells. These experiments demonstrate that combinatorial labeling and super-resolution imaging of single cells is a natural approach to bring systems biology into single cells.
Figures



Comment in
-
Single-cell in situ RNA profiling by sequential hybridization.Nat Methods. 2014 Apr;11(4):360-1. doi: 10.1038/nmeth.2892. Nat Methods. 2014. PMID: 24681720 Free PMC article. No abstract available.
References
-
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–502. - PubMed
-
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5:621–8. - PubMed
-
- Schena M, Shalon D, Davis RW, Brown PO. Quantitative Monitoring of Gene Expression Patterns with a Complementary DNA Microarray. Science. 1995;270:467–470. - PubMed
-
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

