Population structure and linkage disequilibrium in elite barley breeding germplasm from the United States
- PMID: 22661207
- PMCID: PMC3370289
- DOI: 10.1631/jzus.B1200003
Population structure and linkage disequilibrium in elite barley breeding germplasm from the United States
Abstract
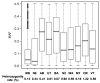
Cultivated barley is known to have a complex population structure and extensive linkage disequilibrium (LD). To conduct robust association mapping (AM) studies of economically important traits in US barley breeding germplasm, population structure and LD decay were examined in a complete panel of US barley breeding germplasm (3840 lines) genotyped with 3072 single nucleotide polymorphisms (SNPs). Nine subpopulations (sp1‒sp9) were identified by the program STRUCTURE and subsequently confirmed by principle component analysis (PCA). Out of the nine subpopulations, seven were very similar to the respective subpopulations identified by Hamblin et al. (2010) which were based on half of the germplasm and half of the SNP markers, but two subpopulations were found to be new. One subpopulation was dominated by six-rowed spring lines from Utah State University (UT) and the other was composed of six-rowed spring lines from multiple breeding programs (USDA-ARS Aberdeen (AB), Busch Agricultural Resources Inc. (BA), UT, and Washington State University (WA)). LD was found to decay across a range from 4.0 to 19.8 cM. This result indicates that the germplasm genotyped with 3072 SNPs would be robust for mapping and possibly identifying the causal polymorphisms contributing to disease resistance and perhaps other traits.
Figures
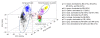
References
-
- Boyko AR, Boyko RH, Boyko CM, Parker HG, Castelhano M, Corey L, Degenhardt JD, Auton A, Hedimbi M, Kityo R. Complex population structure in African village dogs and its implications for inferring dog domestication history. PNAS. 2009;106(33):13903–13908. doi: 10.1073/pnas.0902129106. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

