Lutzomyia umbratilis, the main vector of Leishmania guyanensis, represents a novel species complex?
- PMID: 22662146
- PMCID: PMC3356248
- DOI: 10.1371/journal.pone.0037341
Lutzomyia umbratilis, the main vector of Leishmania guyanensis, represents a novel species complex?
Abstract
Background: Lutzomyia umbratilis is an important Leishmania guyanensis vector in South America. Previous studies have suggested differences in the vector competence between L. umbratilis populations situated on opposite banks of the Amazonas and Negro Rivers in the central Amazonian Brazil region, likely indicating a species complex. However, few studies have been performed on these populations and the taxonomic status of L. umbratilis remains unclear.
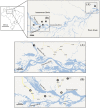
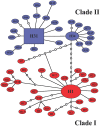
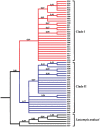
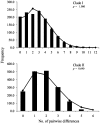
Methodology/principal findings: Phylogeographic structure was estimated for six L. umbratilis samples from the central Amazonian region in Brazil by analyzing mtDNA using 1181 bp of the COI gene to assess whether the populations on opposite banks of these rivers consist of incipient or distinct species. The genetic diversity was fairly high and the results revealed two distinct clades ( = lineages) with 1% sequence divergence. Clade I consisted of four samples from the left bank of the Amazonas and Negro Rivers, whereas clade II comprised two samples from the right bank of Negro River. No haplotypes were shared between samples of two clades. Samples within clades exhibited low to moderate genetic differentiation (F(ST) = -0.0390-0.1841), whereas samples between clades exhibited very high differentiation (F(ST) = 0.7100-0.8497) and fixed differences. These lineages have diverged approximately 0.22 Mya in the middle Pleistocene. Demographic expansion was detected for the lineages I and II approximately 30,448 and 15,859 years ago, respectively, in the late Pleistocene.
Conclusions/significance: The two genetic lineages may represent an advanced speciation stage suggestive of incipient or distinct species within L. umbratilis. These findings suggest that the Amazonas and Negro Rivers may be acting as effective barriers, thus preventing gene flow between populations on opposite sides. Such findings have important implications for epidemiological studies, especially those related to vector competence and anthropophily, and for vector control strategies. In addition, L. umbratilis represents an interesting example in speciation studies.
Conflict of interest statement
Figures
References
-
- Loaiza JR, Bermingham E, Sanjur OI, Scott ME, Bickersmith SA, et al. Review of genetic diversity in malaria vectors (Culicidae: Anophelinae). Inf Genet Evol. 2012;12:1–12. - PubMed
-
- Arrivillaga JC, Norris DE, Feliciangeli MD, Lanzaro GC. Phylogeography of the neotropical sand fly Lutzomyia longipalpis inferred from mitochondrial DNA sequences. Inf Genet Evol. 2002;2:83–95. - PubMed
-
- Bauzer LGCR, Nataly AS, Maingon RDC, Peixoto AA. Lutzomyia longipalpis in Brazil: a complex or a single species? A mini-review. Mem Inst Oswaldo Cruz. 2007;102:1–12. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

