Oligonucleotide microarray analysis of dietary-induced hyperlipidemia gene expression profiles in miniature pigs
- PMID: 22662175
- PMCID: PMC3360772
- DOI: 10.1371/journal.pone.0037581
Oligonucleotide microarray analysis of dietary-induced hyperlipidemia gene expression profiles in miniature pigs
Abstract
Background: Hyperlipidemia animal models have been established, but complete gene expression profiles of the transition from normal lipid levels have not been obtained. Miniature pigs are useful model animals for gene expression studies on dietary-induced hyperlipidemia because they have a similar anatomy and digestive physiology to humans, and blood samples can be obtained from them repeatedly.
Methodology: Two typical dietary treatments were used for dietary-induced hyperlipidemia models, by using specific pathogen-free (SPF) Clawn miniature pigs. One was a high-fat and high-cholesterol diet (HFCD) and the other was a high-fat, high-cholesterol, and high-sucrose diet (HFCSD). Microarray analyses were conducted from whole blood samples during the dietary period and from white blood cells at the end of the dietary period to evaluate the transition of expression profiles of the two dietary models.
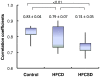
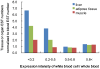
Principal findings: Variations in whole blood gene expression intensity within the HFCD or the HFCSD group were in the same range as the controls provide with normal diet at all periods. This indicates uniformity of dietary-induced hyperlipidemia for our dietary protocols. Gene ontology- (GO) based functional analyses revealed that characteristics of the common changes between HFCD and HFCSD were involved in inflammatory responses and reproduction. The correlation coefficient between whole blood and white blood cell expression profiles at 27 weeks with the HFCSD diet was significantly lower than that of the control and HFCD diet groups. This may be due to the effects of RNA originating from the tissues and/or organs.
Conclusions: No statistically significant differences in fasting plasma lipids and glucose levels between the HFCD and HFCSD groups were observed. However, blood RNA analyses revealed different characteristics corresponding to the dietary protocols. In this study, whole blood RNA analyses proved to be a useful tool to evaluate transitions in dietary-induced hyperlipidemia gene expression profiles in miniature pigs.
Conflict of interest statement
Figures




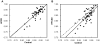
Similar articles
-
Oligonucleotide microarray analysis of age-related gene expression profiles in miniature pigs.PLoS One. 2011;6(5):e19761. doi: 10.1371/journal.pone.0019761. Epub 2011 May 13. PLoS One. 2011. PMID: 21603646 Free PMC article.
-
Differential metabolic and hepatic transcriptome responses of two miniature pig breeds to high dietary cholesterol.Life Sci. 2020 Jun 1;250:117514. doi: 10.1016/j.lfs.2020.117514. Epub 2020 Mar 5. Life Sci. 2020. PMID: 32145306
-
Enteric lactoferrin attenuates the development of high-fat and high-cholesterol diet-induced hypercholesterolemia and atherosclerosis in Microminipigs.Biosci Biotechnol Biochem. 2016;80(2):295-303. doi: 10.1080/09168451.2015.1091713. Epub 2015 Nov 7. Biosci Biotechnol Biochem. 2016. PMID: 26549014
-
Microminipig, a non-rodent experimental animal optimized for life science research:novel atherosclerosis model induced by high fat and cholesterol diet.J Pharmacol Sci. 2011;115(2):115-21. doi: 10.1254/jphs.10r17fm. Epub 2011 Jan 18. J Pharmacol Sci. 2011. PMID: 21258170 Review.
-
Risks for hyperlipidemia.Cardiol Clin. 1986 Feb;4(1):47-56. Cardiol Clin. 1986. PMID: 2871933 Review.
Cited by
-
RNA Sequencing (RNA-Seq) Reveals Extremely Low Levels of Reticulocyte-Derived Globin Gene Transcripts in Peripheral Blood From Horses (Equus caballus) and Cattle (Bos taurus).Front Genet. 2018 Aug 14;9:278. doi: 10.3389/fgene.2018.00278. eCollection 2018. Front Genet. 2018. PMID: 30154823 Free PMC article.
-
Exercise and High-Fat Diet in Obesity: Functional Genomics Perspectives of Two Energy Homeostasis Pillars.Genes (Basel). 2020 Jul 31;11(8):875. doi: 10.3390/genes11080875. Genes (Basel). 2020. PMID: 32752100 Free PMC article. Review.
-
Whole Blood Transcriptomics Is Relevant to Identify Molecular Changes in Response to Genetic Selection for Feed Efficiency and Nutritional Status in the Pig.PLoS One. 2016 Jan 11;11(1):e0146550. doi: 10.1371/journal.pone.0146550. eCollection 2016. PLoS One. 2016. PMID: 26752050 Free PMC article.
-
Hematologic and biochemical reference intervals for specific pathogen free 6-week-old Hampshire-Yorkshire crossbred pigs.J Anim Sci Biotechnol. 2014 Jan 10;5(1):5. doi: 10.1186/2049-1891-5-5. J Anim Sci Biotechnol. 2014. PMID: 24410946 Free PMC article.
References
-
- Lissner L, Heitmann BL. Dietary fat and obesity: evidence from epidemiology. Eur J Clin Nutr. 1995;49(2):79–90. - PubMed
-
- Russell JC, Proctor SD. Small animal models of cardiovascular disease: tools for the study of the roles of metabolic syndrome, dyslipidemia, and atherosclerosis. Cardiovasc Pathol. 2006;15(6):318–330. - PubMed
-
- Oron-Herman M, Kamari Y, Grossman E, Yeger G, Peleg E, et al. Metabolic syndrome: comparison of the two commonly used animal models. Am J Hypertens. 2008;21(9):1018–1022. - PubMed
-
- Kobari Y, Koto M, Tanigawa M. Regression of diet-induced atherosclerosis in Güttingen Miniature Swine. Lab Anim. 1991;25(2):110–116. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

