Nasal bone shape is under complex epistatic genetic control in mouse interspecific recombinant congenic strains
- PMID: 22662199
- PMCID: PMC3360618
- DOI: 10.1371/journal.pone.0037721
Nasal bone shape is under complex epistatic genetic control in mouse interspecific recombinant congenic strains
Abstract
Background: Genetic determinism of cranial morphology in the mouse is still largely unknown, despite the localization of putative QTLs and the identification of genes associated with Mendelian skull malformations. To approach the dissection of this multigenic control, we have used a set of interspecific recombinant congenic strains (IRCS) produced between C57BL/6 and mice of the distant species Mus spretus (SEG/Pas). Each strain has inherited 1.3% of its genome from SEG/Pas under the form of few, small-sized, chromosomal segments.
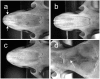
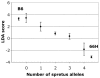
Results: The shape of the nasal bone was studied using outline analysis combined with Fourier descriptors, and differential features were identified between IRCS BcG-66H and C57BL/6. An F2 cross between BcG-66H and C57BL/6 revealed that, out of the three SEG/Pas-derived chromosomal regions present in BcG-66H, two were involved. Segments on chromosomes 1 (∼32 Mb) and 18 (∼13 Mb) showed additive effect on nasal bone shape. The three chromosomal regions present in BcG-66H were isolated in congenic strains to study their individual effect. Epistatic interactions were assessed in bicongenic strains.
Conclusions: Our results show that, besides a strong individual effect, the QTL on chromosome 1 interacts with genes on chromosomes 13 and 18. This study demonstrates that nasal bone shape is under complex genetic control but can be efficiently dissected in the mouse using appropriate genetic tools and shape descriptors.
Conflict of interest statement
Figures





Similar articles
-
Exploration of the genetic organization of morphological modularity on the mouse mandible using a set of interspecific recombinant congenic strains between C57BL/6 and mice of the Mus spretus species.G3 (Bethesda). 2012 Oct;2(10):1257-68. doi: 10.1534/g3.112.003285. Epub 2012 Oct 1. G3 (Bethesda). 2012. PMID: 23050236 Free PMC article.
-
Interspecific recombinant congenic strains between C57BL/6 and mice of the Mus spretus species: a powerful tool to dissect genetic control of complex traits.Genetics. 2007 Dec;177(4):2321-33. doi: 10.1534/genetics.107.078006. Epub 2007 Oct 18. Genetics. 2007. PMID: 17947429 Free PMC article.
-
Genetic analysis of skull shape variation and morphological integration in the mouse using interspecific recombinant congenic strains between C57BL/6 and mice of the mus spretus species.Evolution. 2009 Oct;63(10):2668-86. doi: 10.1111/j.1558-5646.2009.00737.x. Epub 2009 May 28. Evolution. 2009. PMID: 19490077
-
A novel strategy for genetic dissection of complex traits: the population of specific chromosome substitution strains from laboratory and wild mice.Mamm Genome. 2010 Aug;21(7-8):370-6. doi: 10.1007/s00335-010-9270-x. Epub 2010 Jul 11. Mamm Genome. 2010. PMID: 20623355 Review.
-
The AcB/BcA recombinant congenic strains of mice: strategies for phenotype dissection, mapping and cloning of quantitative trait genes.Novartis Found Symp. 2007;281:141-53; discussion 153-5, 208-9. doi: 10.1002/9780470062128.ch12. Novartis Found Symp. 2007. PMID: 17534071 Review.
Cited by
-
Developmental constraint through negative pleiotropy in the zygomatic arch.Evodevo. 2018 Jan 27;9:3. doi: 10.1186/s13227-018-0092-3. eCollection 2018. Evodevo. 2018. PMID: 29423138 Free PMC article.
-
The genetic basis of neurocranial size and shape across varied lab mouse populations.J Anat. 2022 Aug;241(2):211-229. doi: 10.1111/joa.13657. Epub 2022 Mar 31. J Anat. 2022. PMID: 35357006 Free PMC article.
-
Exploration of the genetic organization of morphological modularity on the mouse mandible using a set of interspecific recombinant congenic strains between C57BL/6 and mice of the Mus spretus species.G3 (Bethesda). 2012 Oct;2(10):1257-68. doi: 10.1534/g3.112.003285. Epub 2012 Oct 1. G3 (Bethesda). 2012. PMID: 23050236 Free PMC article.
-
Natural history collections as windows on evolutionary processes.Mol Ecol. 2016 Feb;25(4):864-81. doi: 10.1111/mec.13529. Mol Ecol. 2016. PMID: 26757135 Free PMC article. Review.
References
-
- Leamy LJ, Routman EJ, Cheverud JM. Quantitative trait loci for early- and late-developing skull characters in mice: A test of the genetic independence model of morphological integration. American Naturalist. 1999;153:201–214. - PubMed
-
- Dohmoto A, Shimizu K, Asada Y, Maeda T. Quantitative trait loci on chromosomes 10 and 11 influencing mandible size of SMXA RI mouse strains. J Dent Res. 2002;81:501–504. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

