Maintaining replication origins in the face of genomic change
- PMID: 22665441
- PMCID: PMC3460189
- DOI: 10.1101/gr.138248.112
Maintaining replication origins in the face of genomic change
Abstract
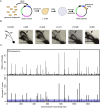
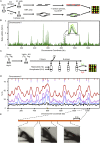
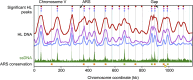
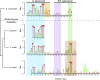
Origins of replication present a paradox to evolutionary biologists. As a collection, they are absolutely essential genomic features, but individually are highly redundant and nonessential. It is therefore difficult to predict to what extent and in what regard origins are conserved over evolutionary time. Here, through a comparative genomic analysis of replication origins and chromosomal replication patterns in the budding yeasts Saccharomyces cerevisiae and Lachancea waltii, we assess to what extent replication origins survived genomic change produced from 150 million years of evolution. We find that L. waltii origins exhibit a core consensus sequence and nucleosome occupancy pattern highly similar to those of S. cerevisiae origins. We further observe that the overall progression of chromosomal replication is similar between L. waltii and S. cerevisiae. Nevertheless, few origins show evidence of being conserved in location between the two species. Among the conserved origins are those surrounding centromeres and adjacent to histone genes, suggesting that proximity to an origin may be important for their regulation. We conclude that, over evolutionary time, origins maintain sequence, structure, and regulation, but are continually being created and destroyed, with the result that their locations are generally not conserved.
Figures
References
-
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 - PubMed
-
- Bailey TL, Gribskov M 1998. Combining evidence using p-values: Application to sequence homology searches. Bioinformatics 14: 48–54 - PubMed
-
- Bell SP 1995. Eukaryotic replicators and associated protein complexes. Curr Opin Genet Dev 5: 162–167 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
