The stress response mediator ATF3 represses androgen signaling by binding the androgen receptor
- PMID: 22665497
- PMCID: PMC3434546
- DOI: 10.1128/MCB.00159-12
The stress response mediator ATF3 represses androgen signaling by binding the androgen receptor
Abstract
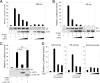
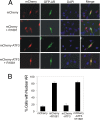
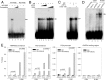
Activating transcription factor 3 (ATF3) is a common mediator of cellular stress response signaling and is often aberrantly expressed in prostate cancer. We report here that ATF3 can directly bind the androgen receptor (AR) and consequently repress AR-mediated gene expression. The ATF3-AR interaction requires the leucine zipper domain of ATF3 that independently binds the DNA-binding and ligand-binding domains of AR, and the interaction prevents AR from binding to cis-acting elements required for expression of androgen-dependent genes while inhibiting the AR N- and C-terminal interaction. The functional consequences of the loss of ATF3 expression include increased transcription of androgen-dependent genes in prostate cancer cells that correlates with increased ability to grow in low-androgen-containing medium and increased proliferative activity of the prostate epithelium in ATF3 knockout mice that is associated with prostatic hyperplasia. Our results thus demonstrate that ATF3 is a novel repressor of androgen signaling that can inhibit AR functions, allowing prostate cells to restore homeostasis and maintain integrity in the face of a broad spectrum of intrinsic and environmental insults.
Figures





Similar articles
-
Botanical Formulation HX109 Ameliorates TP-Induced Benign Prostate Hyperplasia in Rat Model and Inhibits Androgen Receptor Signaling by Upregulating Ca2+/CaMKKβ and ATF3 in LNCaP Cells.Nutrients. 2018 Dec 7;10(12):1946. doi: 10.3390/nu10121946. Nutrients. 2018. PMID: 30544543 Free PMC article.
-
Edelfosine Promotes Apoptosis in Androgen-Deprived Prostate Tumors by Increasing ATF3 and Inhibiting Androgen Receptor Activity.Mol Cancer Ther. 2016 Jun;15(6):1353-63. doi: 10.1158/1535-7163.MCT-15-0332. Epub 2016 Mar 4. Mol Cancer Ther. 2016. PMID: 26944919 Free PMC article.
-
Revealing the mechanism of ethyl acetate extracts of Semen Impatientis against prostate cancer based on network pharmacology and transcriptomics.J Ethnopharmacol. 2024 Aug 10;330:118228. doi: 10.1016/j.jep.2024.118228. Epub 2024 Apr 19. J Ethnopharmacol. 2024. PMID: 38643863
-
Androgen-selective gene regulation in the prostate.Biomed Pharmacother. 2002 Mar;56(2):78-83. doi: 10.1016/s0753-3322(01)00159-7. Biomed Pharmacother. 2002. PMID: 12000138 Review.
-
Androgen action in the prostate gland.Minerva Urol Nefrol. 2012 Mar;64(1):35-49. Minerva Urol Nefrol. 2012. PMID: 22402316 Review.
Cited by
-
ATF3 deficiency impairs the proliferative-secretory phase transition and decidualization in RIF patients.Cell Death Dis. 2021 Apr 12;12(4):387. doi: 10.1038/s41419-021-03679-8. Cell Death Dis. 2021. PMID: 33846304 Free PMC article.
-
Emerging roles of ATF3 in the suppression of prostate cancer.Mol Cell Oncol. 2015 Feb 3;3(1):e1010948. doi: 10.1080/23723556.2015.1010948. eCollection 2016 Jan. Mol Cell Oncol. 2015. PMID: 27308526 Free PMC article.
-
Transient receptor potential vanilloid subtype 1 depletion mediates mechanical allodynia through cellular signal alterations in small-fiber neuropathy.Pain Rep. 2021 Apr 2;6(1):e922. doi: 10.1097/PR9.0000000000000922. eCollection 2021. Pain Rep. 2021. PMID: 34585035 Free PMC article. Review.
-
ONC201 Targets AR and AR-V7 Signaling, Reduces PSA, and Synergizes with Everolimus in Prostate Cancer.Mol Cancer Res. 2018 May;16(5):754-766. doi: 10.1158/1541-7786.MCR-17-0614. Epub 2018 Mar 27. Mol Cancer Res. 2018. PMID: 29588330 Free PMC article.
-
Botanical Formulation HX109 Ameliorates TP-Induced Benign Prostate Hyperplasia in Rat Model and Inhibits Androgen Receptor Signaling by Upregulating Ca2+/CaMKKβ and ATF3 in LNCaP Cells.Nutrients. 2018 Dec 7;10(12):1946. doi: 10.3390/nu10121946. Nutrients. 2018. PMID: 30544543 Free PMC article.
References
-
- Adachi M, et al. 2000. Androgen-insensitivity syndrome as a possible coactivator disease. N. Engl. J. Med. 343:856–862 - PubMed
-
- Andersen R, et al. 2010. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell 17:535–546 - PubMed
-
- Beleza-Meireles A, et al. 2008. Activating transcription 3: a hormone responsive gene in the etiology of hypospadias. Eur. J. Endocrinol. 158:729–739 - PubMed
-
- Brinkmann AO. 2001. Molecular basis of androgen insensitivity. Mol. Cell Endocrinol. 179:105–109 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
